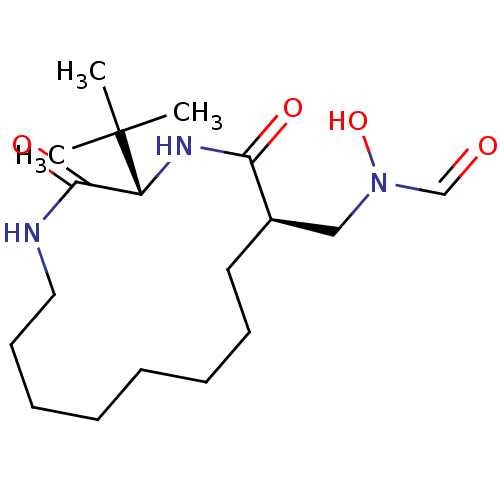
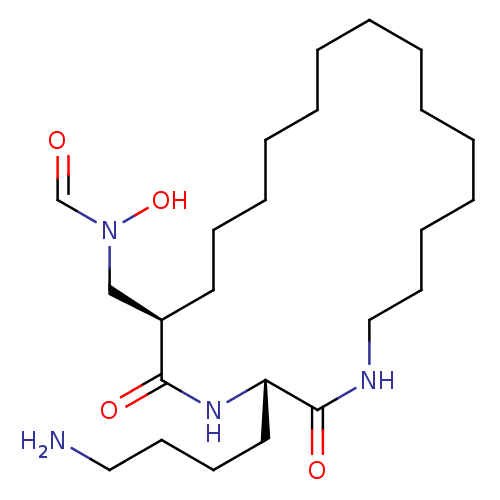
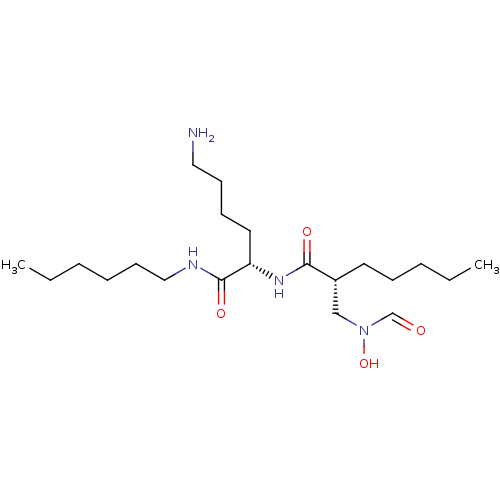
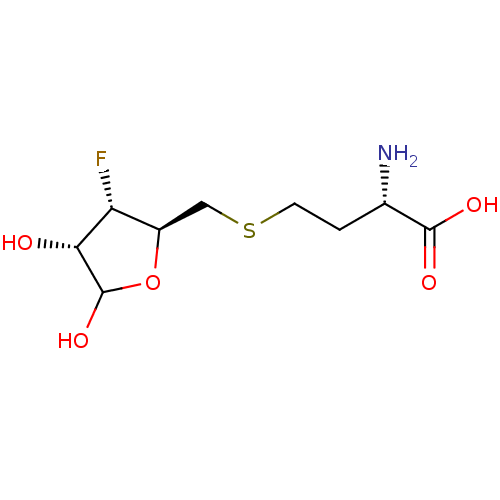
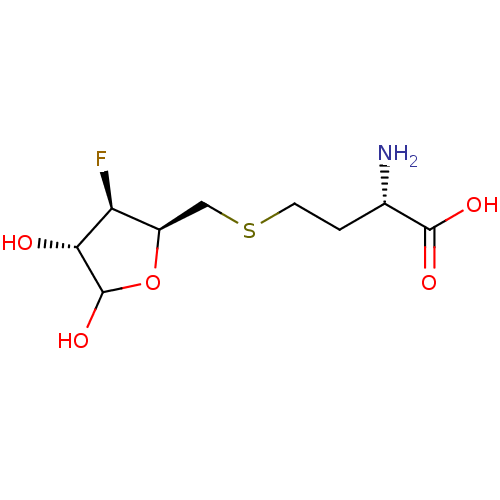
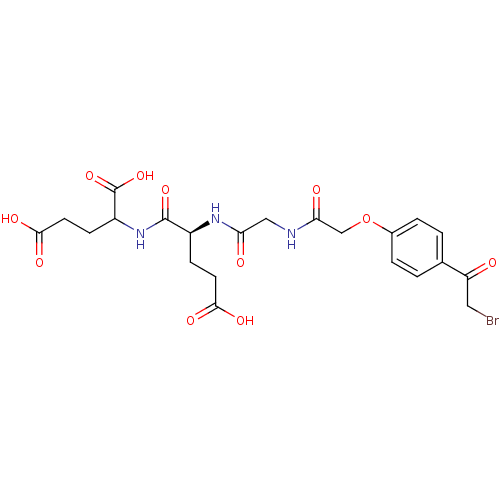
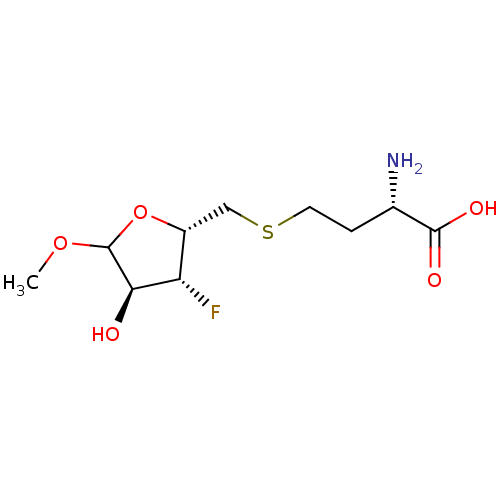
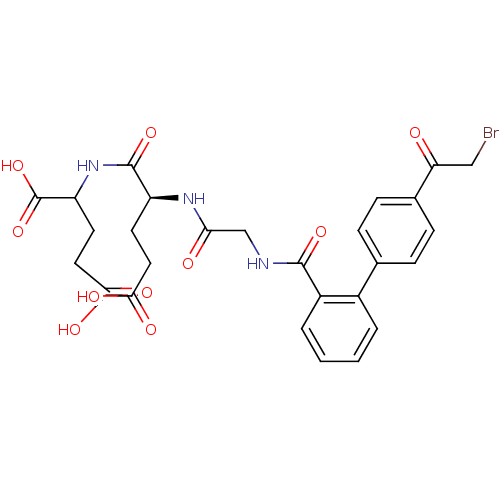
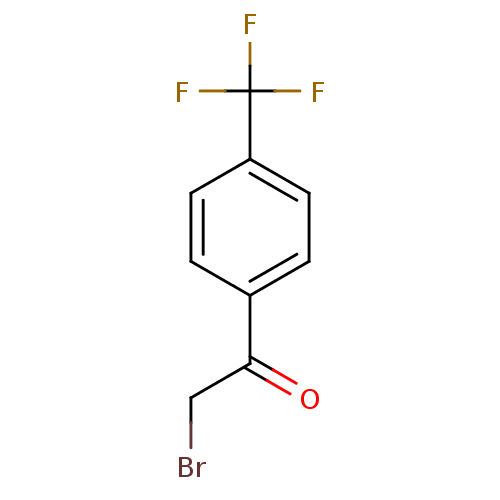
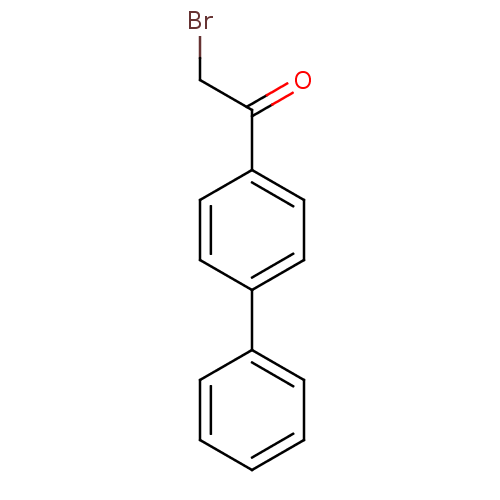
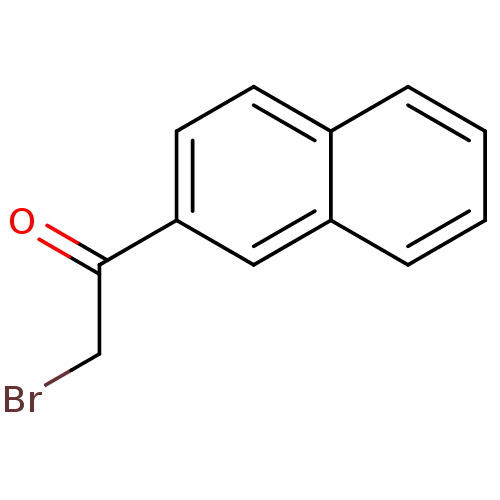
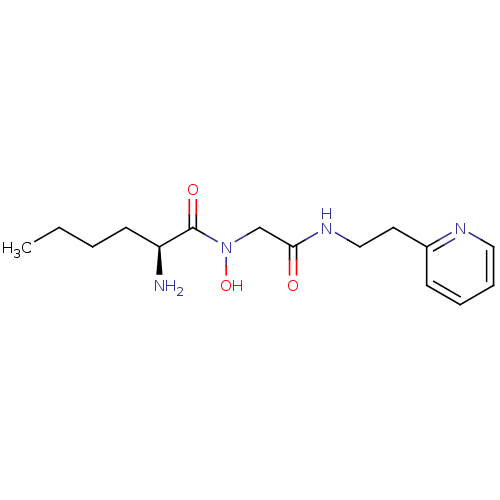
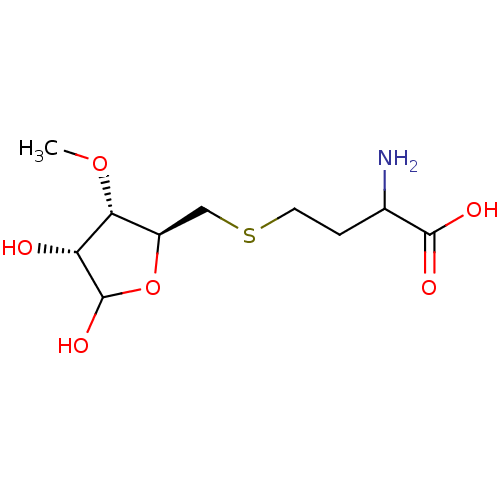
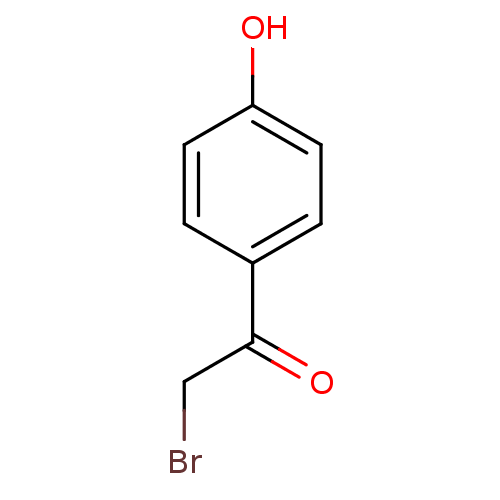
Affinity DataKi: 0.220nMAssay Description:Inhibitory effect against E. coli peptide deformylase (PDF) by AAP assayMore data for this Ligand-Target Pair
Affinity DataKi: 0.230nMAssay Description:Inhibitory effect against E. coli peptide deformylase (PDF) by AAP assayMore data for this Ligand-Target Pair
Affinity DataKi: 0.330nMAssay Description:Inhibitory effect against E. coli peptide deformylase (PDF) by AAP assayMore data for this Ligand-Target Pair
Affinity DataKi: 0.670nMAssay Description:Binding affinity against Co(II)-substituted E. coli peptide deformylase was determinedMore data for this Ligand-Target Pair
Affinity DataKi: 3nMAssay Description:Inhibitory effect against E. coli peptide deformylase (PDF) by AAP assayMore data for this Ligand-Target Pair
Affinity DataKi: 11nMAssay Description:Inhibitory effect against E. coli peptide deformylase (PDF) by DPPI assayMore data for this Ligand-Target Pair
Affinity DataKi: 12nMAssay Description:Inhibitory effect against E. coli peptide deformylase (PDF) by AAP assayMore data for this Ligand-Target Pair
Affinity DataKi: 13.7nMAssay Description:Inhibitory effect against E. coli peptide deformylase (PDF) by AAP assayMore data for this Ligand-Target Pair
Affinity DataKi: 23nMAssay Description:Inhibitory effect against E. coli peptide deformylase (PDF) by DPPI assayMore data for this Ligand-Target Pair
Affinity DataKi: 25nMAssay Description:Inhibitory effect against E. coli peptide deformylase (PDF) by AAP assayMore data for this Ligand-Target Pair
Affinity DataKi: 36nMAssay Description:Inhibitory effect against E. coli peptide deformylase (PDF) by AAP assayMore data for this Ligand-Target Pair
Affinity DataKi: 63nMAssay Description:Inhibitory effect against E. coli peptide deformylase (PDF) by DPPI assayMore data for this Ligand-Target Pair
Affinity DataKi: 74nMAssay Description:Inhibitory effect against E. coli peptide deformylase (PDF) by DPPI assayMore data for this Ligand-Target Pair
Affinity DataKi: 77nMAssay Description:Inhibitory effect against E. coli peptide deformylase (PDF) by DPPI assayMore data for this Ligand-Target Pair
Affinity DataKi: 92nMAssay Description:Inhibitory effect against E. coli peptide deformylase (PDF) by DPPI assayMore data for this Ligand-Target Pair
Affinity DataKi: 96nMAssay Description:Inhibitory effect against E. coli peptide deformylase (PDF) by DPPI assayMore data for this Ligand-Target Pair
Affinity DataKi: 109nMAssay Description:Inhibitory effect against E. coli peptide deformylase (PDF) by DPPI assayMore data for this Ligand-Target Pair
Affinity DataKi: 258nMAssay Description:Inhibitory effect against E. coli peptide deformylase (PDF) by DPPI assayMore data for this Ligand-Target Pair
Affinity DataKi: 370nMAssay Description:Inhibition of cobalt substituted Bacillus subtilis LuxSMore data for this Ligand-Target Pair
Affinity DataKi: 430nMAssay Description:Inhibition of ferrous substituted Bacillus subtilis LuxSMore data for this Ligand-Target Pair
Affinity DataKi: 430nMAssay Description:Inhibition of Bacillus subtilis LuxS assessed as enzyme-inhibitor dissociation constantMore data for this Ligand-Target Pair
Affinity DataKi: 700nMAssay Description:Inhibition of Bacillus subtilis LuxS assessed as enzyme-inhibitor dissociation constantMore data for this Ligand-Target Pair
Affinity DataKi: 710nMAssay Description:Inhibitory effect against E. coli peptide deformylase (PDF) by DPPI assayMore data for this Ligand-Target Pair
Affinity DataKi: 720nMAssay Description:Inhibition of ferrous substituted Bacillus subtilis LuxSMore data for this Ligand-Target Pair
Affinity DataKi: 720nMAssay Description:Inhibition of cobalt substituted Bacillus subtilis LuxSMore data for this Ligand-Target Pair
Affinity DataKi: 1.40E+3nMAssay Description:Inhibition of Bacillus subtilis LuxS assessed as enzyme-inhibitor dissociation constantMore data for this Ligand-Target Pair
Affinity DataKi: 2.10E+3nMAssay Description:Inhibitory effect against E. coli peptide deformylase (PDF) by DPPI assayMore data for this Ligand-Target Pair
Affinity DataKi: 2.80E+3nMAssay Description:Inhibition of Bacillus subtilis LuxS assessed as enzyme-inhibitor dissociation constantMore data for this Ligand-Target Pair
TargetTyrosine-protein phosphatase non-receptor type 1(Homo sapiens (Human))
The Ohio State University
Curated by ChEMBL
The Ohio State University
Curated by ChEMBL
Affinity DataKi: 2.80E+3nMAssay Description:Dissociation constant of the compound towards Protein-tyrosine phosphatase 1B receptor-inhibitor complex was determined using PNP as substrateMore data for this Ligand-Target Pair
TargetS-ribosylhomocysteine lyase(Escherichia coli (strain K12))
The Ohio State University
Curated by ChEMBL
The Ohio State University
Curated by ChEMBL
Affinity DataKi: 3.20E+3nMAssay Description:Inhibition of cobalt substituted Escherichia coli LuxSMore data for this Ligand-Target Pair
Affinity DataKi: 4.20E+3nMAssay Description:Inhibition of Bacillus subtilis LuxSMore data for this Ligand-Target Pair
Affinity DataKi: 7.70E+3nMAssay Description:Inhibition of Bacillus subtilis LuxSMore data for this Ligand-Target Pair
Affinity DataKi: 7.90E+3nMAssay Description:Inhibition of Bacillus subtilis LuxSMore data for this Ligand-Target Pair
Affinity DataKi: 8.80E+3nMAssay Description:Inhibition of Bacillus subtilis LuxS assessed as enzyme-inhibitor dissociation constantMore data for this Ligand-Target Pair
TargetS-ribosylhomocysteine lyase(Vibrio harveyi (strain ATCC BAA-1116 / BB120))
The Ohio State University
Curated by ChEMBL
The Ohio State University
Curated by ChEMBL
Affinity DataKi: 9.70E+3nMAssay Description:Inhibition of cobalt substituted Vibrio harveyi LuxSMore data for this Ligand-Target Pair
TargetTyrosine-protein phosphatase non-receptor type 1(Homo sapiens (Human))
The Ohio State University
Curated by ChEMBL
The Ohio State University
Curated by ChEMBL
Affinity DataKi: 9.90E+3nMAssay Description:Dissociation constant of the compound towards Protein-tyrosine phosphatase 1B receptor-inhibitor complex was determined using PNP as substrateMore data for this Ligand-Target Pair
Affinity DataKi: 1.06E+4nMAssay Description:Inhibition of Bacillus subtilis LuxSMore data for this Ligand-Target Pair
Affinity DataKi: 1.06E+4nMAssay Description:Inhibition of zinc substituted Bacillus subtilis LuxSMore data for this Ligand-Target Pair
TargetS-ribosylhomocysteine lyase(Escherichia coli (strain K12))
The Ohio State University
Curated by ChEMBL
The Ohio State University
Curated by ChEMBL
Affinity DataKi: 1.27E+4nMAssay Description:Inhibition of cobalt substituted Escherichia coli LuxSMore data for this Ligand-Target Pair
TargetS-ribosylhomocysteine lyase(Vibrio harveyi (strain ATCC BAA-1116 / BB120))
The Ohio State University
Curated by ChEMBL
The Ohio State University
Curated by ChEMBL
Affinity DataKi: 1.28E+4nMAssay Description:Inhibition of cobalt substituted Vibrio harveyi LuxSMore data for this Ligand-Target Pair
TargetTyrosine-protein phosphatase non-receptor type 1(Homo sapiens (Human))
The Ohio State University
Curated by ChEMBL
The Ohio State University
Curated by ChEMBL
Affinity DataKi: 1.40E+4nMAssay Description:Dissociation constant of the compound towards Protein-tyrosine phosphatase 1B receptor-inhibitor complex was determined using PNP as substrateMore data for this Ligand-Target Pair
TargetTyrosine-protein phosphatase non-receptor type 1(Homo sapiens (Human))
The Ohio State University
Curated by ChEMBL
The Ohio State University
Curated by ChEMBL
Affinity DataKi: 1.40E+4nMAssay Description:Dissociation constant of the compound towards Protein-tyrosine phosphatase 1B receptor-inhibitor complex was determined using PNP as substrateMore data for this Ligand-Target Pair
Affinity DataKi: 1.96E+4nMAssay Description:Inhibition of zinc substituted Bacillus subtilis LuxSMore data for this Ligand-Target Pair
TargetTyrosine-protein phosphatase non-receptor type 1(Homo sapiens (Human))
The Ohio State University
Curated by ChEMBL
The Ohio State University
Curated by ChEMBL
Affinity DataKi: 2.50E+4nMAssay Description:Dissociation constant of the compound towards Protein-tyrosine phosphatase 1B receptor-inhibitor complex was determined using PNP as substrateMore data for this Ligand-Target Pair
TargetTyrosine-protein phosphatase non-receptor type 6(Homo sapiens (Human))
The Ohio State University
Curated by ChEMBL
The Ohio State University
Curated by ChEMBL
Affinity DataKi: 2.90E+4nMAssay Description:Dissociation constant of the compound towards SHP 1 receptor-inhibitor complex was determined using PNP as substrateMore data for this Ligand-Target Pair
Affinity DataKi: 3.30E+4nMAssay Description:Inhibitory activity of the compound against Methionine aminopeptidase 1More data for this Ligand-Target Pair
Affinity DataKi: 3.70E+4nMAssay Description:Compound was evaluated for inhibitory activity against Peptide deformylaseMore data for this Ligand-Target Pair
Affinity DataKi: 4.20E+4nMAssay Description:Inhibition of Bacillus subtilis LuxSMore data for this Ligand-Target Pair
Affinity DataKi: 4.20E+4nMAssay Description:Inhibition of Bacillus subtilis LuxSMore data for this Ligand-Target Pair
TargetTyrosine-protein phosphatase non-receptor type 1(Homo sapiens (Human))
The Ohio State University
Curated by ChEMBL
The Ohio State University
Curated by ChEMBL
Affinity DataKi: 4.30E+4nMAssay Description:Dissociation constant of the compound towards Protein-tyrosine phosphatase 1B receptor-inhibitor complex was determined using PNP as substrateMore data for this Ligand-Target Pair

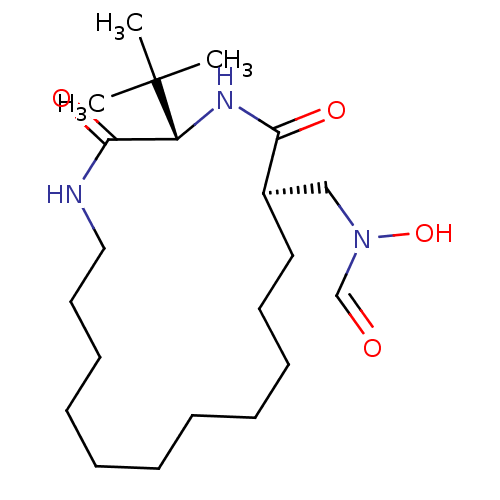


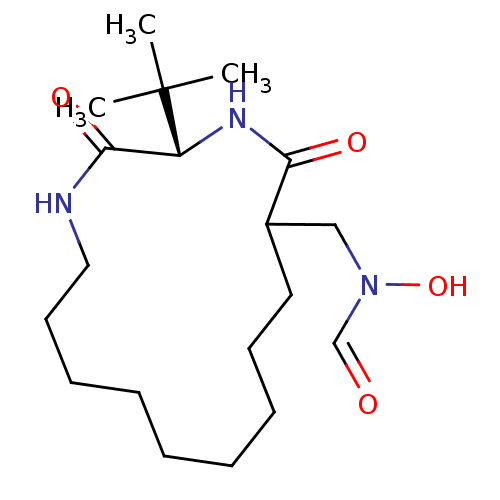

 3D Structure (crystal)
3D Structure (crystal)