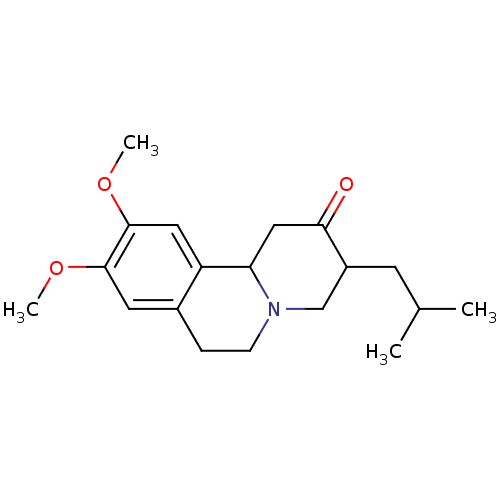
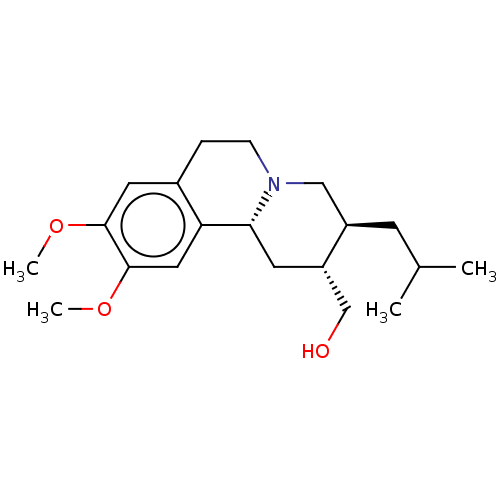
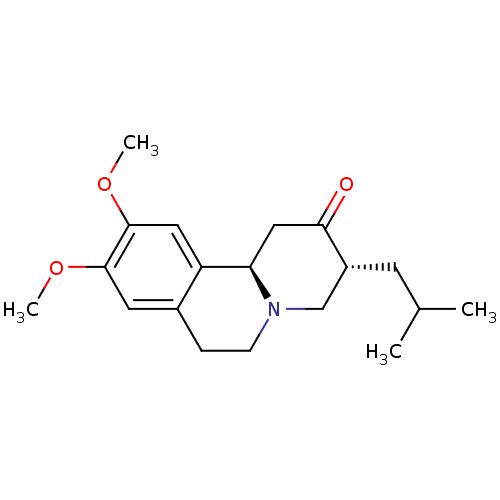
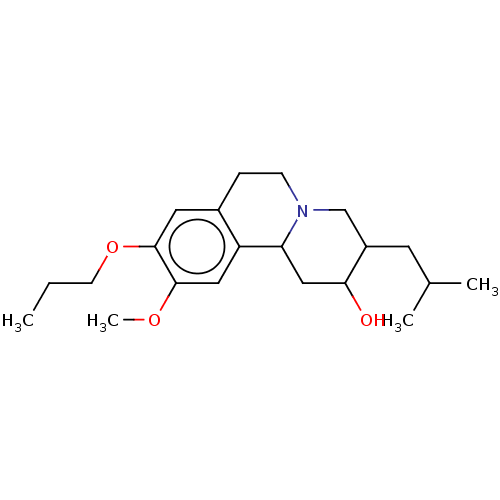
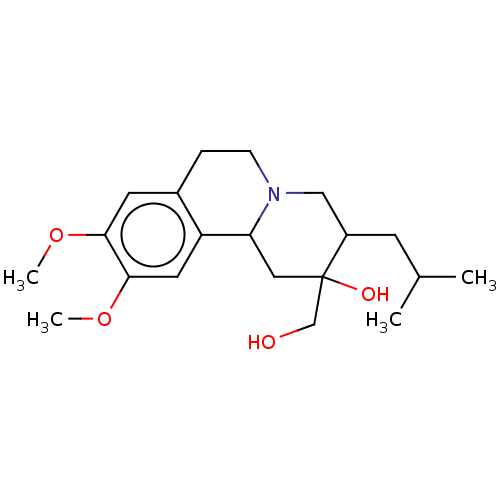
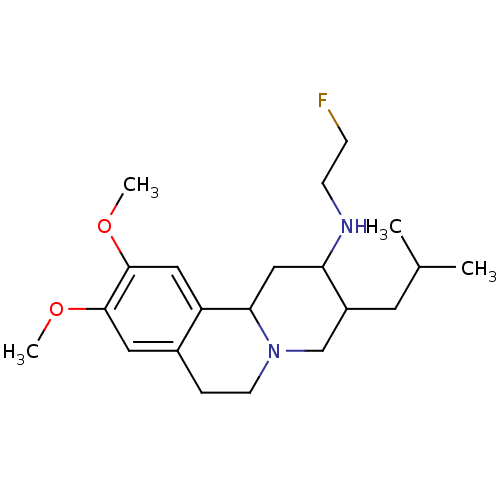
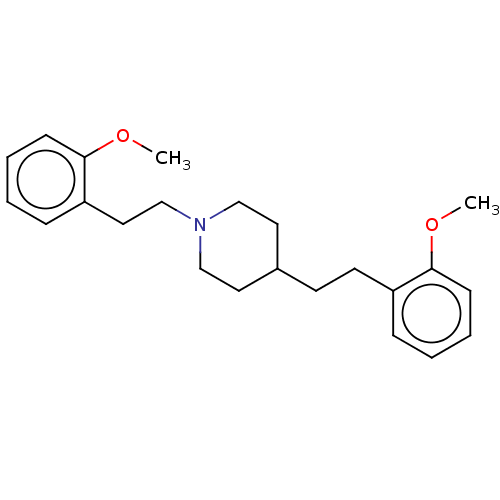
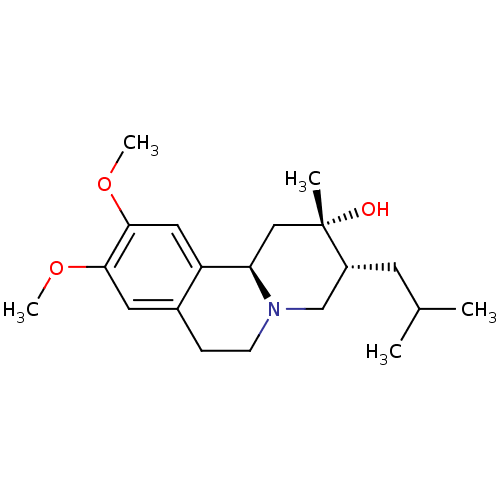
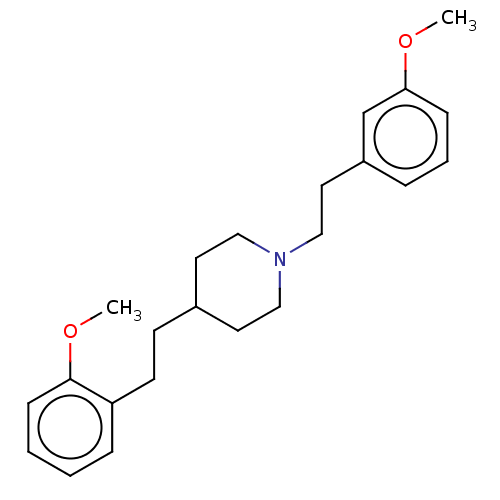
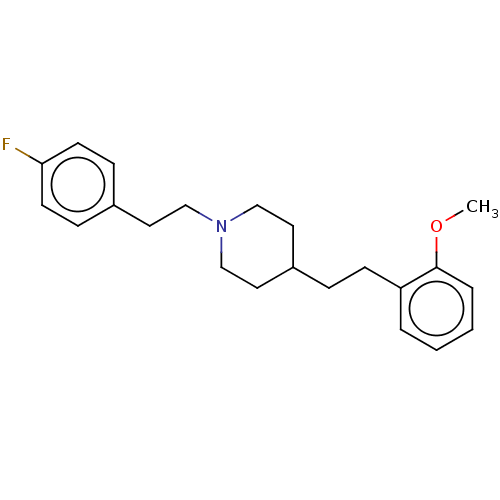Affinity DataKi: 0.100nMAssay Description:Displacement of [125I]-iodovinyl-TBZ from VMAT2 in rat striatal homogenate after 60 mins by gamma countingMore data for this Ligand-Target Pair
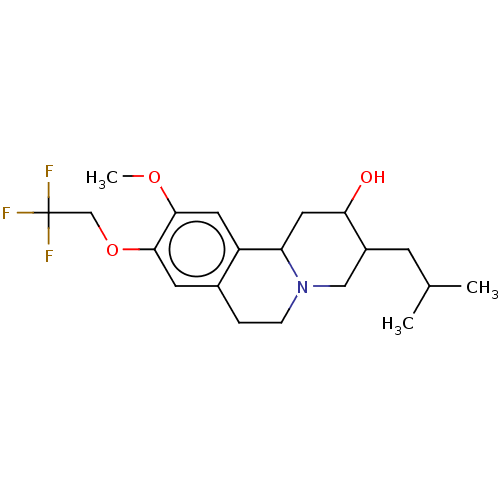
Affinity DataKi: 0.240nMAssay Description:Displacement of [125I]-iodovinyl-TBZ from VMAT2 in rat striatal membrane homogenatesMore data for this Ligand-Target Pair
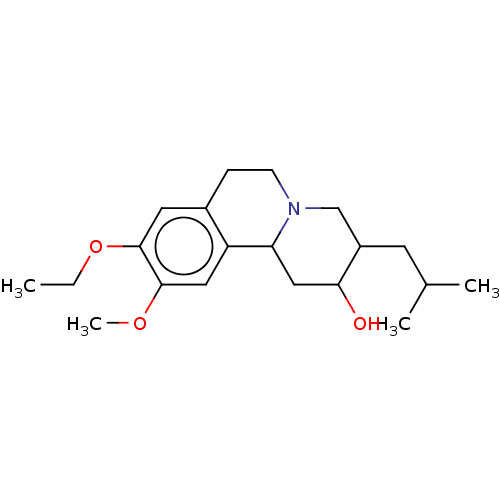
Affinity DataKi: 0.360nMAssay Description:Displacement of [3H]-DTBZ from VMAT2 in rat striatal membrane homogenatesMore data for this Ligand-Target Pair
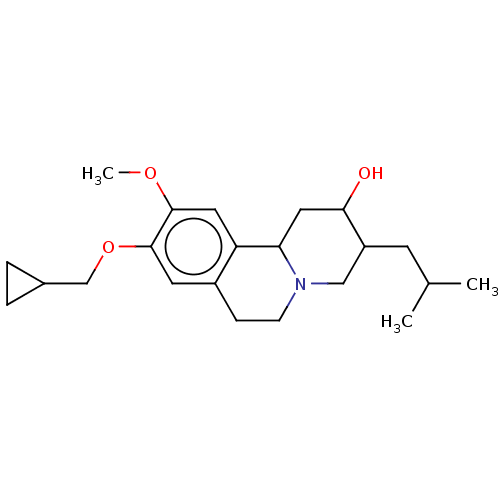
Affinity DataKi: 1.03nMAssay Description:Displacement of [125I]-iodovinyl-TBZ from VMAT2 in rat striatal homogenate after 60 mins by gamma countingMore data for this Ligand-Target Pair
Affinity DataIC50: 1.30nMpH: 8.5Assay Description:The uptake assay was performed in reaction buffer containing 140 mM K2-tartrate, 10 mM Tricine, 10 mM Tris, and 5 mM MgCl2, pH 8.5. Liposomes (1 _...More data for this Ligand-Target Pair
Affinity DataKi: 1.90nMAssay Description:Examples of techniques for determining the capability of a compound to inhibit VMAT2 are provided below. The procedure is adapted from that described...More data for this Ligand-Target Pair
Affinity DataKi: 1.90nMAssay Description:Examples of techniques for determining the capability of a compound to inhibit VMAT2 are provided below. The procedure is adapted from that described...More data for this Ligand-Target Pair
Affinity DataKi: 1.90nMAssay Description:Examples of techniques for determining the capability of a compound to inhibit VMAT2 are provided below. The procedure is adapted from that described...More data for this Ligand-Target Pair
Affinity DataIC50: 14.5nM Kd: 2nMpH: 7.5 T: 2°CAssay Description:Liposomes (1-2 ul) were added to 200 ul of reaction buffer containing 150 mM NaCl, 15 mM Tris-HCl, pH 7.5, and increasing concentrations of [3H]TBZOH...More data for this Ligand-Target Pair
Affinity DataKi: 2nMAssay Description:Inhibition of VMAT2 in rat brainMore data for this Ligand-Target Pair
Affinity DataKi: 2nMAssay Description:Inhibition of VMAT2 in rat brainMore data for this Ligand-Target Pair
Affinity DataIC50: 2nMpH: 8.5Assay Description:The uptake assay was performed in reaction buffer containing 140 mM K2-tartrate, 10 mM Tricine, 10 mM Tris, and 5 mM MgCl2, pH 8.5. Liposomes (1 _...More data for this Ligand-Target Pair
Affinity DataKi: 2.45nMAssay Description:Displacement of [125I]-iodovinyl-TBZ from VMAT2 in rat striatal membrane homogenatesMore data for this Ligand-Target Pair
Affinity DataIC50: 2.5nMpH: 8.5Assay Description:The uptake assay was performed in reaction buffer containing 140 mM K2-tartrate, 10 mM Tricine, 10 mM Tris, and 5 mM MgCl2, pH 8.5. Liposomes (1 _...More data for this Ligand-Target Pair
Affinity DataKi: 2.60nMAssay Description:In vitro inhibition of [3H]- Methoxytetrabenazine binding to rat brain vesicular monoamine transporter 2 (VMAT2)More data for this Ligand-Target Pair
Affinity DataKi: 2.60nMAssay Description:Examples of techniques for determining the capability of a compound to inhibit VMAT2 are provided below. The procedure is adapted from that described...More data for this Ligand-Target Pair
Affinity DataIC50: 3.70nMpH: 8.5Assay Description:The uptake assay was performed in reaction buffer containing 140 mM K2-tartrate, 10 mM Tricine, 10 mM Tris, and 5 mM MgCl2, pH 8.5. Liposomes (1 _...More data for this Ligand-Target Pair
Affinity DataKi: 3.75nMAssay Description:Displacement of [3H]-DTBZ from VMAT2 in rat striatal membrane homogenatesMore data for this Ligand-Target Pair
Affinity DataIC50: 3.80nMpH: 8.5Assay Description:The uptake assay was performed in reaction buffer containing 140 mM K2-tartrate, 10 mM Tricine, 10 mM Tris, and 5 mM MgCl2, pH 8.5. Liposomes (1 _...More data for this Ligand-Target Pair
Affinity DataKi: 3.96nMAssay Description:Displacement of [3H]DHTBZ from Sprague-Dawley rat striatum VMAT2 after 1 hr by liquid scintillation countingMore data for this Ligand-Target Pair
Affinity DataKi: 4nMAssay Description:Inhibition constant against [3H]nicotine binding to vesicular monoamine transporter-2 of rat brain membranesMore data for this Ligand-Target Pair
Affinity DataKd: 4nMpH: 7.5 T: 2°CAssay Description:Liposomes (1-2 ul) were added to 200 ul of reaction buffer containing 150 mM NaCl, 15 mM Tris-HCl, pH 7.5, and increasing concentrations of [3H]TBZOH...More data for this Ligand-Target Pair
Affinity DataKi: 4.20nMAssay Description:Inhibition of [3H]-DHTBZ binding to VMAT2 from rat forebrain membranesMore data for this Ligand-Target Pair
Affinity DataIC50: 4.30nMpH: 8.5Assay Description:The uptake assay was performed in reaction buffer containing 140 mM K2-tartrate, 10 mM Tricine, 10 mM Tris, and 5 mM MgCl2, pH 8.5. Liposomes (1 _...More data for this Ligand-Target Pair
Affinity DataKi: 4.47nMAssay Description:Displacement of [3H]DHTBZ from Sprague-Dawley rat striatum VMAT2 after 1 hr by liquid scintillation countingMore data for this Ligand-Target Pair
Affinity DataIC50: 4.5nMpH: 8.5Assay Description:The uptake assay was performed in reaction buffer containing 140 mM K2-tartrate, 10 mM Tricine, 10 mM Tris, and 5 mM MgCl2, pH 8.5. Liposomes (1 _...More data for this Ligand-Target Pair
Affinity DataIC50: 4.90nMAssay Description:Inhibition of [3H]DTBZ binding to VMAT2 in rat brain incubated for 30 mins by liquid scintillation spectrometry analysisMore data for this Ligand-Target Pair
Affinity DataIC50: 4.90nMpH: 8.5Assay Description:The uptake assay was performed in reaction buffer containing 140 mM K2-tartrate, 10 mM Tricine, 10 mM Tris, and 5 mM MgCl2, pH 8.5. Liposomes (1 _...More data for this Ligand-Target Pair
Affinity DataIC50: 5.10nMAssay Description:Inhibition of [3H]DA uptake at VMAT2 in rat striatal synaptosome incubated for 10 mins followed by [3H]DA addition and measured after 20 mins by TopC...More data for this Ligand-Target Pair
Affinity DataIC50: 5.10nMAssay Description:Inhibition of [3H]DTBZ binding to VMAT2 in rat brain incubated for 30 mins by liquid scintillation spectrometry analysisMore data for this Ligand-Target Pair
Affinity DataIC50: 8.30nM Kd: 5.20nMpH: 7.5 T: 2°CAssay Description:Liposomes (1-2 ul) were added to 200 ul of reaction buffer containing 150 mM NaCl, 15 mM Tris-HCl, pH 7.5, and increasing concentrations of [3H]TBZOH...More data for this Ligand-Target Pair
Affinity DataIC50: 5.70nMAssay Description:Inhibition of [3H]DA uptake at VMAT2 in rat striatal synaptosome incubated for 10 mins followed by [3H]DA addition and measured after 20 mins by TopC...More data for this Ligand-Target Pair
Affinity DataKi: 5.98nMAssay Description:Displacement of [125I]-iodovinyl-TBZ from VMAT2 in rat striatal homogenate after 60 mins by gamma countingMore data for this Ligand-Target Pair
Affinity DataIC50: 6nMAssay Description:Inhibition of [3H]DTBZ binding to VMAT2 in rat brain incubated for 30 mins by liquid scintillation spectrometry analysisMore data for this Ligand-Target Pair
Affinity DataIC50: 6nMAssay Description:Inhibition of [3H]DA uptake at VMAT2 in rat striatal synaptosome incubated for 10 mins followed by [3H]DA addition and measured after 20 mins by TopC...More data for this Ligand-Target Pair
Affinity DataKi: 7.01nMAssay Description:Displacement of [3H]-DTBZ from VMAT2 in rat striatal membrane homogenatesMore data for this Ligand-Target Pair
Affinity DataKi: 7.62nMAssay Description:Displacement of [3H]DHTBZ from Sprague-Dawley rat striatum VMAT2 after 1 hr by liquid scintillation countingMore data for this Ligand-Target Pair
Affinity DataIC50: 8nMAssay Description:Inhibition of [3H]DTBZ binding to VMAT2 in rat brain incubated for 30 mins by liquid scintillation spectrometry analysisMore data for this Ligand-Target Pair
Affinity DataKi: 8.10nMAssay Description:In vitro inhibition of [3H]- Methoxytetrabenazine binding to rat brain vesicular monoamine transporter 2 (VMAT2)More data for this Ligand-Target Pair
Affinity DataKi: 8.63nMAssay Description:Displacement of [3H]-DTBZ from VMAT2 in rat striatal membrane homogenatesMore data for this Ligand-Target Pair
Affinity DataKi: 8.68nMAssay Description:Displacement of [125I]-iodovinyl-TBZ from VMAT2 in rat striatal membrane homogenatesMore data for this Ligand-Target Pair
Affinity DataKi: 9.21nMAssay Description:Displacement of [125I]-iodovinyl-TBZ from VMAT2 in rat striatal membrane homogenatesMore data for this Ligand-Target Pair
Affinity DataKi: 9.30nMAssay Description:Inhibition of [3H]dopamine uptake at VMAT2 in rat brain synaptic vesicle by liquid scintillation spectroscopyMore data for this Ligand-Target Pair
Affinity DataKi: 9.30nMAssay Description:Inhibition of [3H]DA uptake at VMAT2 in rat striata vesicles homogenate after 8 mins by liquid scintillation countingMore data for this Ligand-Target Pair
Affinity DataKi: 9.70nMAssay Description:Inhibition of VMAT2 in rat brainMore data for this Ligand-Target Pair
Affinity DataIC50: 11nMAssay Description:Inhibition of [3H]DA uptake at VMAT2 in rat striatal synaptosome incubated for 10 mins followed by [3H]DA addition and measured after 20 mins by TopC...More data for this Ligand-Target Pair
Affinity DataKi: 12nMAssay Description:In vitro inhibition of [3H]- Methoxytetrabenazine binding to rat brain vesicular monoamine transporter 2 (VMAT2)More data for this Ligand-Target Pair
Affinity DataIC50: 13nMAssay Description:Inhibition of [3H]DTBZ binding to VMAT2 in rat brain incubated for 30 mins by liquid scintillation spectrometry analysisMore data for this Ligand-Target Pair
Affinity DataKi: 13nMAssay Description:Inhibition of [3H]DA uptake at VMAT2 in rat striata vesicles homogenate after 8 mins by liquid scintillation countingMore data for this Ligand-Target Pair
Affinity DataKi: 13nMAssay Description:Inhibition of [3H]DA uptake at VMAT2 in rat striata vesicles homogenate after 8 mins by liquid scintillation countingMore data for this Ligand-Target Pair
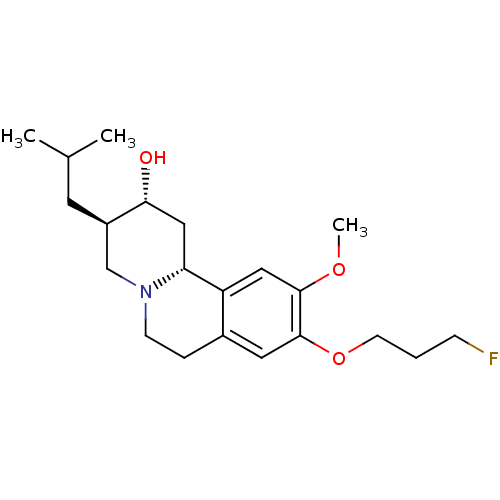
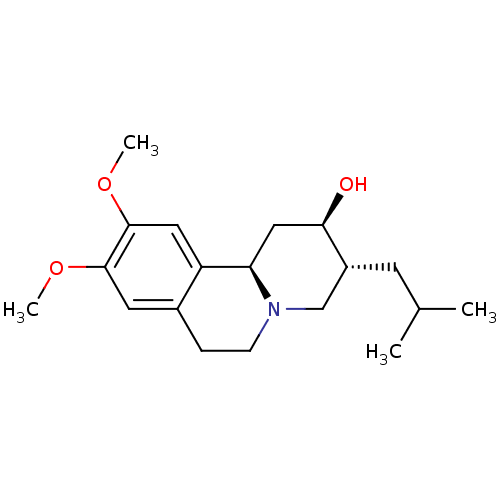
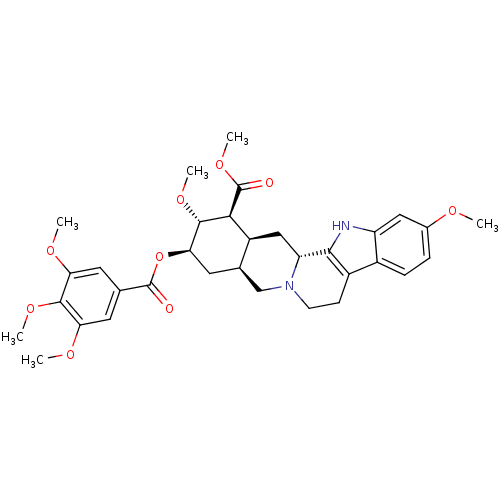
 3D Structure (crystal)
3D Structure (crystal)