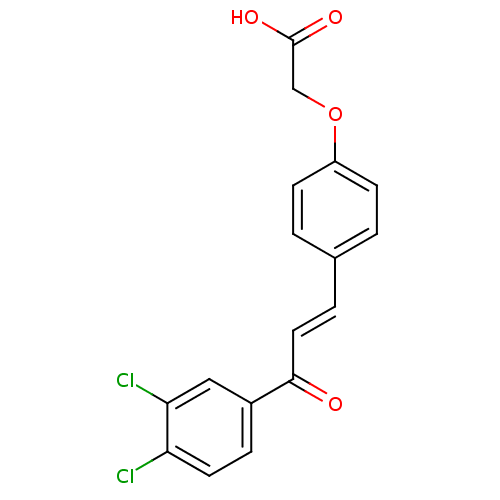
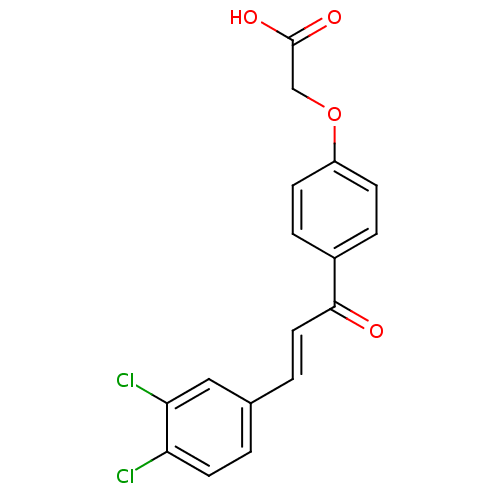
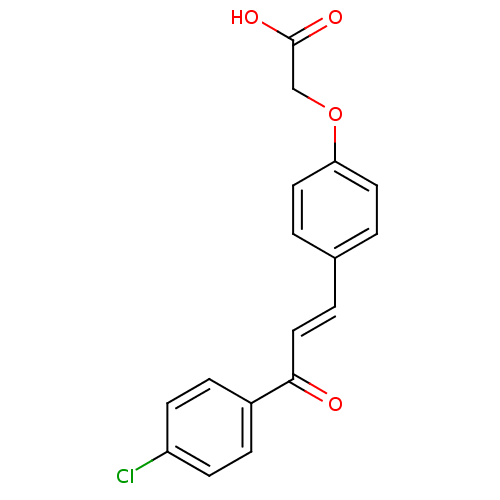
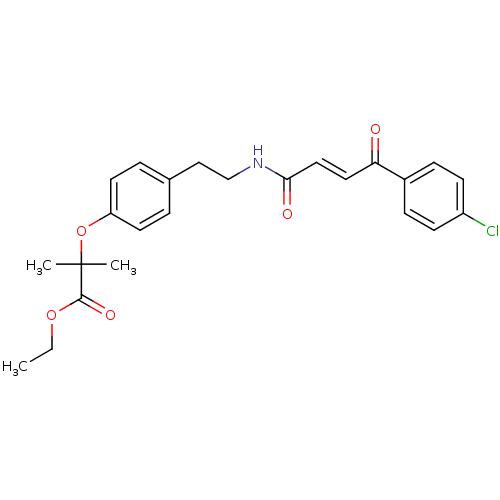
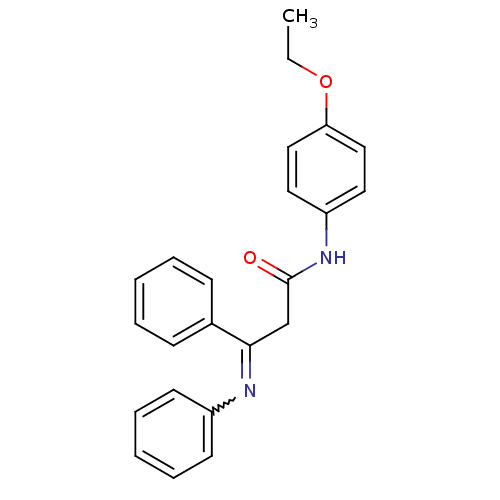
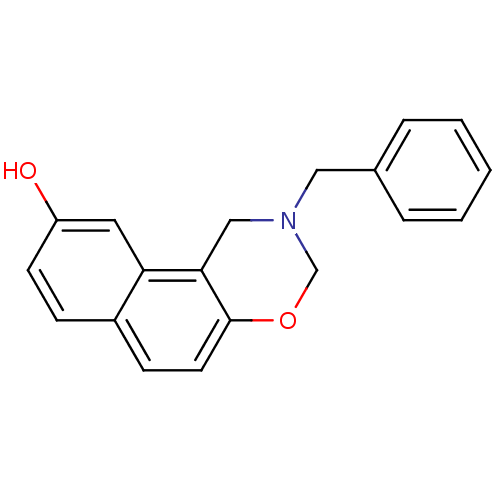
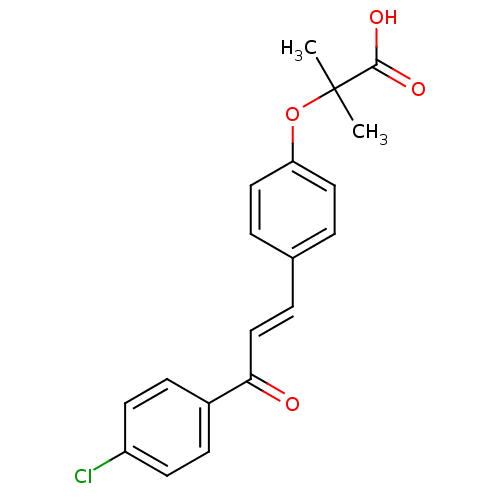
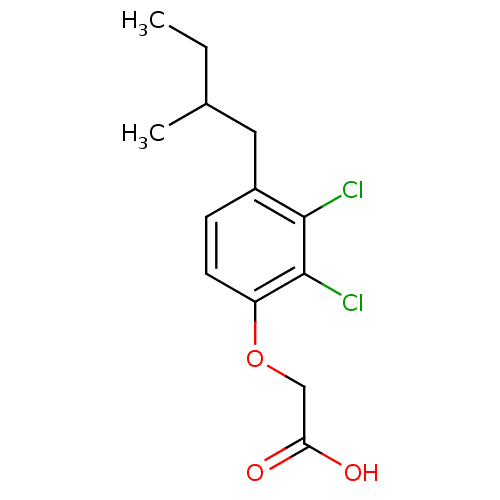
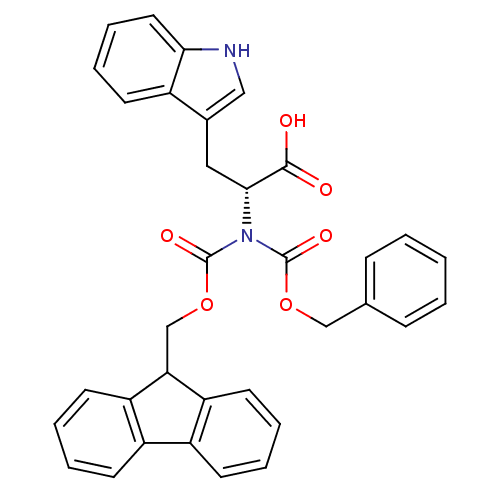
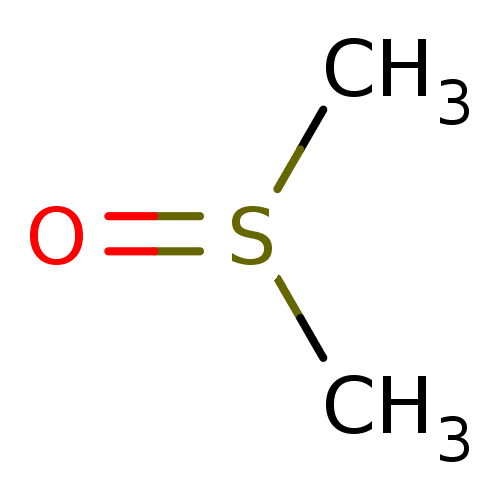Affinity DataIC50: 4.90E+4nMpH: 7.4 T: 2°CAssay Description:Interference of the p53-MDM2 binding by test compounds was measured in a 96-well polypropylene round-bottom microtiter plate (Costar, Serocluster). H...More data for this Ligand-Target Pair
Affinity DataIC50: 1.17E+5nMpH: 7.4 T: 2°CAssay Description:Interference of the p53-MDM2 binding by test compounds was measured in a 96-well polypropylene round-bottom microtiter plate (Costar, Serocluster). H...More data for this Ligand-Target Pair
Affinity DataIC50: 2.06E+5nMpH: 7.4 T: 2°CAssay Description:Interference of the p53-MDM2 binding by test compounds was measured in a 96-well polypropylene round-bottom microtiter plate (Costar, Serocluster). H...More data for this Ligand-Target Pair
Affinity DataIC50: >2.50E+5nMpH: 7.4 T: 2°CAssay Description:Interference of the p53-MDM2 binding by test compounds was measured in a 96-well polypropylene round-bottom microtiter plate (Costar, Serocluster). H...More data for this Ligand-Target Pair
Affinity DataIC50: >2.50E+5nMpH: 7.4 T: 2°CAssay Description:Interference of the p53-MDM2 binding by test compounds was measured in a 96-well polypropylene round-bottom microtiter plate (Costar, Serocluster). H...More data for this Ligand-Target Pair
Affinity DataIC50: >2.50E+5nMpH: 7.4 T: 2°CAssay Description:Interference of the p53-MDM2 binding by test compounds was measured in a 96-well polypropylene round-bottom microtiter plate (Costar, Serocluster). H...More data for this Ligand-Target Pair
Affinity DataIC50: >2.50E+5nMpH: 7.4 T: 2°CAssay Description:Interference of the p53-MDM2 binding by test compounds was measured in a 96-well polypropylene round-bottom microtiter plate (Costar, Serocluster). H...More data for this Ligand-Target Pair
Affinity DataIC50: 2.50E+5nMpH: 7.4 T: 2°CAssay Description:Interference of the p53-MDM2 binding by test compounds was measured in a 96-well polypropylene round-bottom microtiter plate (Costar, Serocluster). H...More data for this Ligand-Target Pair
Affinity DataIC50: >2.50E+5nMpH: 7.4 T: 2°CAssay Description:Interference of the p53-MDM2 binding by test compounds was measured in a 96-well polypropylene round-bottom microtiter plate (Costar, Serocluster). H...More data for this Ligand-Target Pair
TargetInsulin-like growth factor-binding protein 5(Homo sapiens (Human))
Institute For Biochemistry
Curated by ChEMBL
Institute For Biochemistry
Curated by ChEMBL
Affinity DataKd: 1.64E+6nMAssay Description:Dissociation constant of the compound in DMSO binding to Insulin-like growth factor binding protein 5 at the residue R87More data for this Ligand-Target Pair
TargetInsulin-like growth factor-binding protein 5(Homo sapiens (Human))
Institute For Biochemistry
Curated by ChEMBL
Institute For Biochemistry
Curated by ChEMBL
Affinity DataKd: 2.42E+6nMAssay Description:Dissociation constant of the compound in DMSO binding to Insulin-like growth factor binding protein 5 at the residue K91More data for this Ligand-Target Pair
TargetInsulin-like growth factor-binding protein 5(Homo sapiens (Human))
Institute For Biochemistry
Curated by ChEMBL
Institute For Biochemistry
Curated by ChEMBL
Affinity DataKd: 2.78E+6nMAssay Description:Dissociation constant of the compound binding to Insulin-like growth factor binding protein 5 at the residue L-81More data for this Ligand-Target Pair
TargetInsulin-like growth factor-binding protein 5(Homo sapiens (Human))
Institute For Biochemistry
Curated by ChEMBL
Institute For Biochemistry
Curated by ChEMBL
Affinity DataKd: 1.38E+6nMAssay Description:Dissociation constant of the compound in DMSO binding to Insulin-like growth factor binding protein 5 at the residue S85More data for this Ligand-Target Pair
TargetInsulin-like growth factor-binding protein 5(Homo sapiens (Human))
Institute For Biochemistry
Curated by ChEMBL
Institute For Biochemistry
Curated by ChEMBL
Affinity DataKd: 7.83E+8nMAssay Description:Dissociation constant of the compound in binding to Insulin-like growth factor binding protein 5 at the residue Y86More data for this Ligand-Target Pair
TargetInsulin-like growth factor-binding protein 5(Homo sapiens (Human))
Institute For Biochemistry
Curated by ChEMBL
Institute For Biochemistry
Curated by ChEMBL
Affinity DataKd: 1.09E+6nMAssay Description:Dissociation constant of the compound binding to Insulin-like growth factor binding protein 5 at the residue L-81More data for this Ligand-Target Pair
TargetInsulin-like growth factor-binding protein 5(Homo sapiens (Human))
Institute For Biochemistry
Curated by ChEMBL
Institute For Biochemistry
Curated by ChEMBL
Affinity DataKd: 2.36E+6nMAssay Description:Dissociation constant of the compound in PBS binding to Insulin-like growth factor binding protein 5 at the residue R87More data for this Ligand-Target Pair
TargetInsulin-like growth factor-binding protein 5(Homo sapiens (Human))
Institute For Biochemistry
Curated by ChEMBL
Institute For Biochemistry
Curated by ChEMBL
Affinity DataKd: 6.10E+8nMAssay Description:Dissociation constant of the compound in binding to Insulin-like growth factor binding protein 5 at the residue L81More data for this Ligand-Target Pair
TargetInsulin-like growth factor-binding protein 5(Homo sapiens (Human))
Institute For Biochemistry
Curated by ChEMBL
Institute For Biochemistry
Curated by ChEMBL
Affinity DataKd: 2.33E+6nMAssay Description:Dissociation constant of the compound in PBS binding to Insulin-like growth factor binding protein 5 at the residue S85More data for this Ligand-Target Pair
TargetInsulin-like growth factor-binding protein 5(Homo sapiens (Human))
Institute For Biochemistry
Curated by ChEMBL
Institute For Biochemistry
Curated by ChEMBL
Affinity DataKd: 2.93E+6nMAssay Description:Dissociation constant of the compound in PBS binding to Insulin-like growth factor binding protein 5 at the residue L73More data for this Ligand-Target Pair
TargetInsulin-like growth factor-binding protein 5(Homo sapiens (Human))
Institute For Biochemistry
Curated by ChEMBL
Institute For Biochemistry
Curated by ChEMBL
Affinity DataKd: 1.58E+6nMAssay Description:Dissociation constant of the compound in DMSO binding to Insulin-like growth factor binding protein 5 at the residue Y50More data for this Ligand-Target Pair
TargetInsulin-like growth factor-binding protein 5(Homo sapiens (Human))
Institute For Biochemistry
Curated by ChEMBL
Institute For Biochemistry
Curated by ChEMBL
Affinity DataKd: 1.08E+6nMAssay Description:Dissociation constant of the compound binding to Insulin-like growth factor binding protein 5 at the residue L-81More data for this Ligand-Target Pair
TargetInsulin-like growth factor-binding protein 5(Homo sapiens (Human))
Institute For Biochemistry
Curated by ChEMBL
Institute For Biochemistry
Curated by ChEMBL
Affinity DataKd: 2.78E+6nMAssay Description:Dissociation constant of the compound in DMSO binding to Insulin-like growth factor binding protein 5 at the residue L81More data for this Ligand-Target Pair
TargetInsulin-like growth factor-binding protein 5(Homo sapiens (Human))
Institute For Biochemistry
Curated by ChEMBL
Institute For Biochemistry
Curated by ChEMBL
Affinity DataKd: 7.19E+8nMAssay Description:Dissociation constant of the compound in BS binding to Insulin-like growth factor binding protein 5 at the residue K91More data for this Ligand-Target Pair
TargetInsulin-like growth factor-binding protein 5(Homo sapiens (Human))
Institute For Biochemistry
Curated by ChEMBL
Institute For Biochemistry
Curated by ChEMBL
Affinity DataKd: 6.50E+8nMAssay Description:Dissociation constant of the compound in binding to Insulin-like growth factor binding protein 5 at the residue S85More data for this Ligand-Target Pair
TargetInsulin-like growth factor-binding protein 5(Homo sapiens (Human))
Institute For Biochemistry
Curated by ChEMBL
Institute For Biochemistry
Curated by ChEMBL
Affinity DataKd: 2.12E+6nMAssay Description:Dissociation constant of the compound in PBS binding to Insulin-like growth factor binding protein 5 at the residue K91More data for this Ligand-Target Pair
TargetInsulin-like growth factor-binding protein 5(Homo sapiens (Human))
Institute For Biochemistry
Curated by ChEMBL
Institute For Biochemistry
Curated by ChEMBL
Affinity DataKd: 5.41E+8nMAssay Description:Dissociation constant of the compound in binding to Insulin-like growth factor binding protein 5 at the residue L73More data for this Ligand-Target Pair
TargetInsulin-like growth factor-binding protein 5(Homo sapiens (Human))
Institute For Biochemistry
Curated by ChEMBL
Institute For Biochemistry
Curated by ChEMBL
Affinity DataKd: 1.72E+6nMAssay Description:Dissociation constant of the compound in PBS binding to Insulin-like growth factor binding protein 5 at the residue Y86More data for this Ligand-Target Pair
TargetInsulin-like growth factor-binding protein 5(Homo sapiens (Human))
Institute For Biochemistry
Curated by ChEMBL
Institute For Biochemistry
Curated by ChEMBL
Affinity DataKd: 6.48E+8nMAssay Description:Dissociation constant of the compound in binding to Insulin-like growth factor binding protein 5 at the residue Y50More data for this Ligand-Target Pair
TargetInsulin-like growth factor-binding protein 5(Homo sapiens (Human))
Institute For Biochemistry
Curated by ChEMBL
Institute For Biochemistry
Curated by ChEMBL
Affinity DataKd: 2.88E+6nMAssay Description:Dissociation constant of the compound in PBS binding to Insulin-like growth factor binding protein 5 at the residue L81More data for this Ligand-Target Pair
TargetInsulin-like growth factor-binding protein 5(Homo sapiens (Human))
Institute For Biochemistry
Curated by ChEMBL
Institute For Biochemistry
Curated by ChEMBL
Affinity DataKd: 1.31E+6nMAssay Description:Dissociation constant of the compound in DMSO binding to Insulin-like growth factor binding protein 5 at the residue L73More data for this Ligand-Target Pair
TargetInsulin-like growth factor-binding protein 5(Homo sapiens (Human))
Institute For Biochemistry
Curated by ChEMBL
Institute For Biochemistry
Curated by ChEMBL
Affinity DataKd: 1.82E+6nMAssay Description:Dissociation constant of the compound in PBS binding to Insulin-like growth factor binding protein 5 at the residue Y50More data for this Ligand-Target Pair
TargetInsulin-like growth factor-binding protein 5(Homo sapiens (Human))
Institute For Biochemistry
Curated by ChEMBL
Institute For Biochemistry
Curated by ChEMBL
Affinity DataKd: 1.90E+6nMAssay Description:Dissociation constant of the compound in DMSO binding to Insulin-like growth factor binding protein 5 at the residue Y86More data for this Ligand-Target Pair
TargetInsulin-like growth factor-binding protein 5(Homo sapiens (Human))
Institute For Biochemistry
Curated by ChEMBL
Institute For Biochemistry
Curated by ChEMBL
Affinity DataKd: 9.21E+8nMAssay Description:Dissociation constant of the compound in binding to Insulin-like growth factor binding protein 5 at the residue R87More data for this Ligand-Target Pair
TargetInsulin-like growth factor-binding protein 5(Homo sapiens (Human))
Institute For Biochemistry
Curated by ChEMBL
Institute For Biochemistry
Curated by ChEMBL
Affinity DataKd: 7.18E+5nMAssay Description:Dissociation constant of the compound binding to Insulin-like growth factor binding protein 5 at the residue L-81More data for this Ligand-Target Pair
Affinity DataKd: 2.70E+5nMpH: 7.4 T: 2°CAssay Description:NMR measurements consisted of monitoring changes in chemical shifts and line widths of the backbone amide resonances of uniformly 15N-enriched MDM2 s...More data for this Ligand-Target Pair
Affinity DataKd: 2.50E+5nMpH: 7.4 T: 2°CAssay Description:NMR measurements consisted of monitoring changes in chemical shifts and line widths of the backbone amide resonances of uniformly 15N-enriched MDM2 s...More data for this Ligand-Target Pair
Affinity DataKd: 2.44E+5nMpH: 7.4 T: 2°CAssay Description:NMR measurements consisted of monitoring changes in chemical shifts and line widths of the backbone amide resonances of uniformly 15N-enriched MDM2 s...More data for this Ligand-Target Pair
Affinity DataKd: 1.50E+5nMpH: 7.4 T: 2°CAssay Description:NMR measurements consisted of monitoring changes in chemical shifts and line widths of the backbone amide resonances of uniformly 15N-enriched MDM2 s...More data for this Ligand-Target Pair
Affinity DataKd: 9.00E+4nMpH: 7.4 T: 2°CAssay Description:NMR measurements consisted of monitoring changes in chemical shifts and line widths of the backbone amide resonances of uniformly 15N-enriched MDM2 s...More data for this Ligand-Target Pair
Affinity DataKd: 2.20E+5nMpH: 7.4 T: 2°CAssay Description:NMR measurements consisted of monitoring changes in chemical shifts and line widths of the backbone amide resonances of uniformly 15N-enriched MDM2 s...More data for this Ligand-Target Pair
TargetInsulin-like growth factor-binding protein 5(Homo sapiens (Human))
Institute For Biochemistry
Curated by ChEMBL
Institute For Biochemistry
Curated by ChEMBL
Affinity DataKd: 4.32E+4nMAssay Description:Dissociation constant of the compound binding to Insulin-like growth factor binding protein 5 at the residue L-81More data for this Ligand-Target Pair