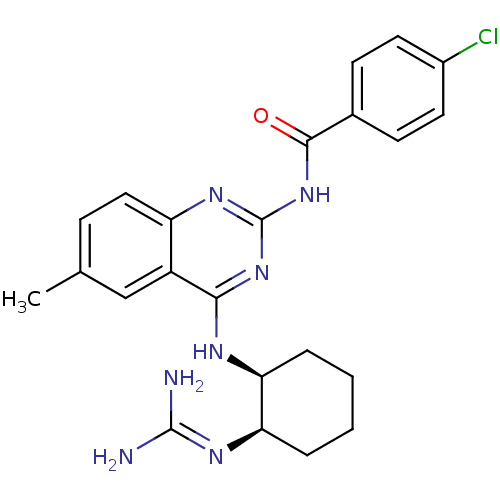
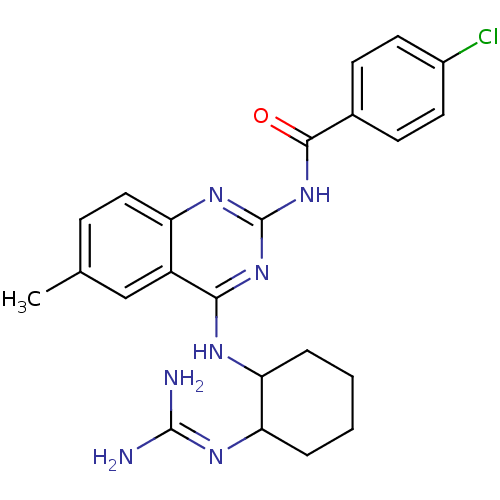
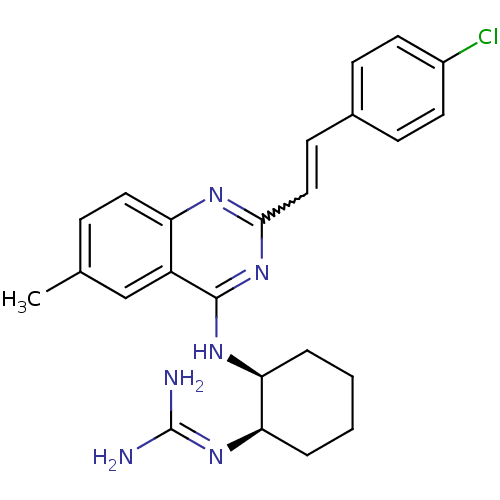
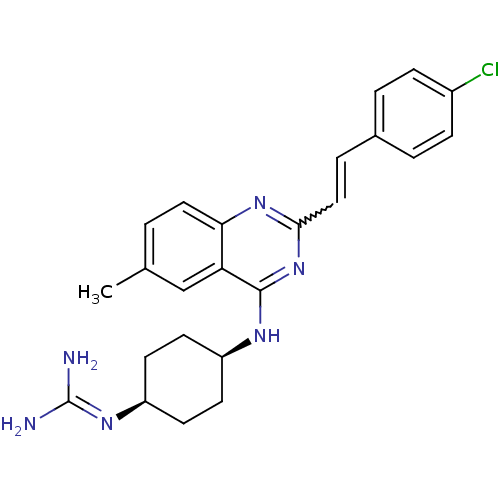
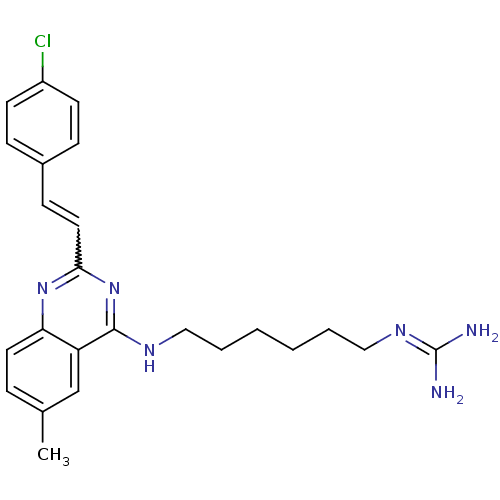
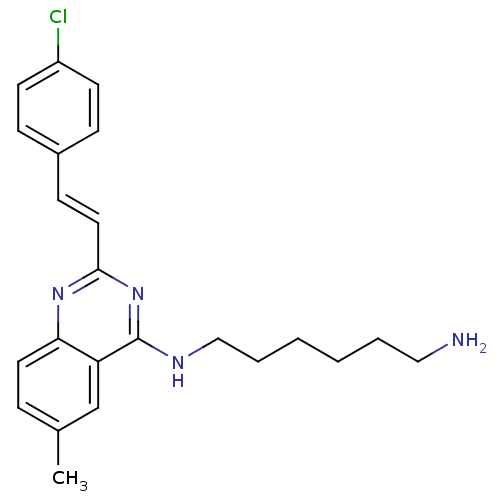
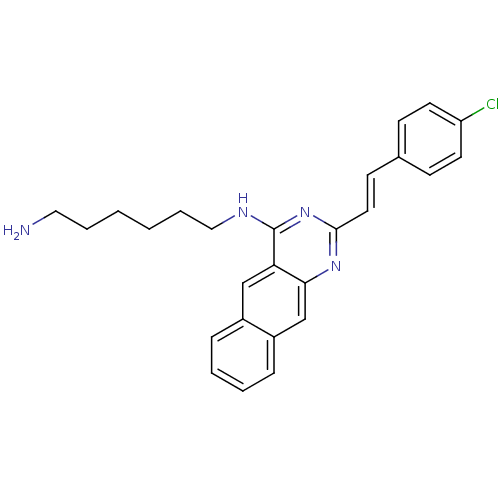
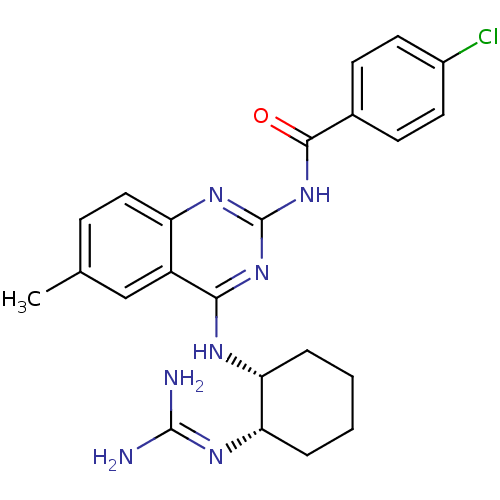
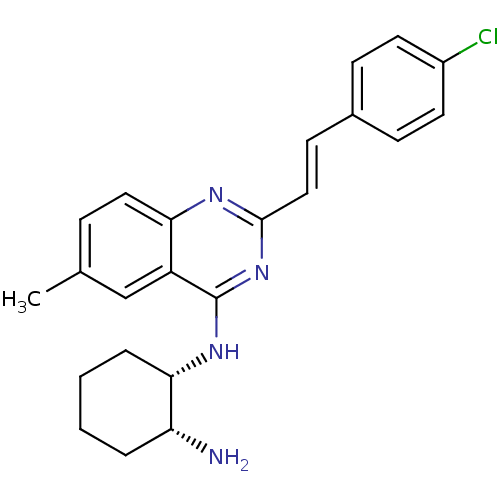
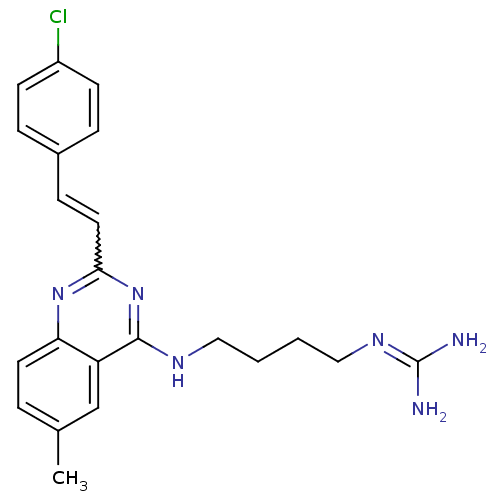
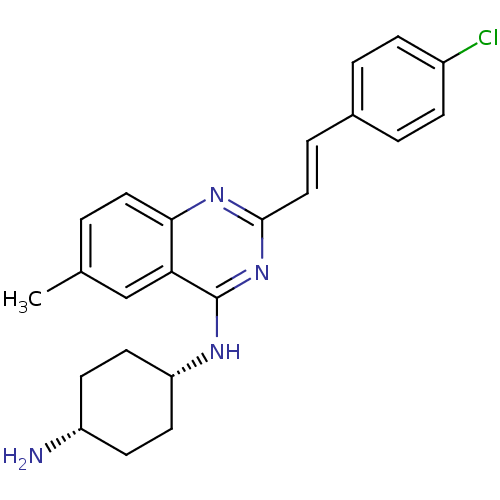
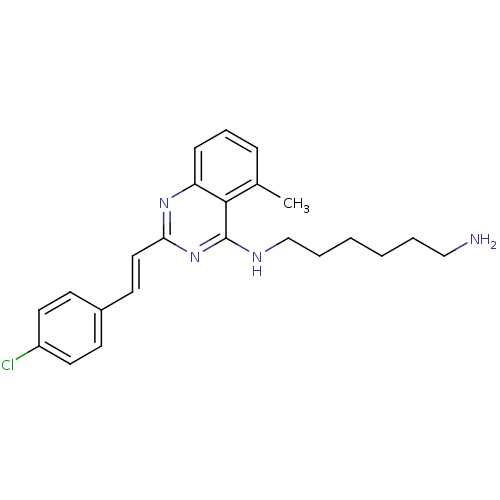
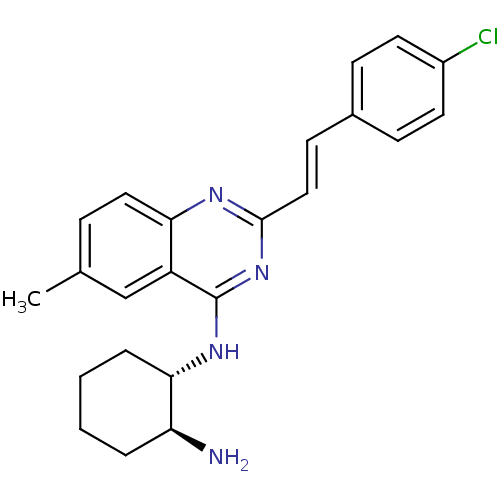
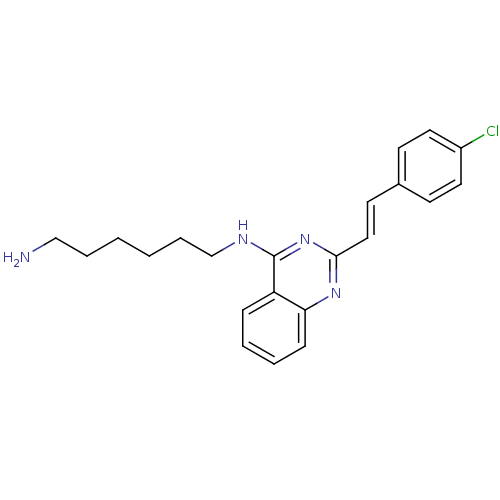
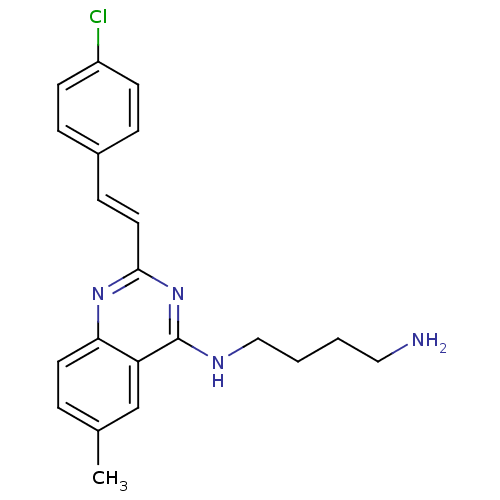
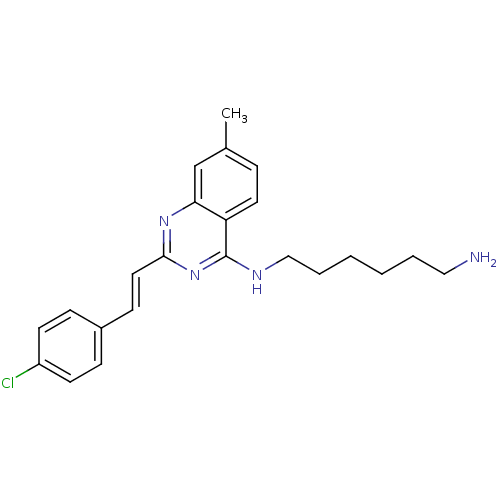
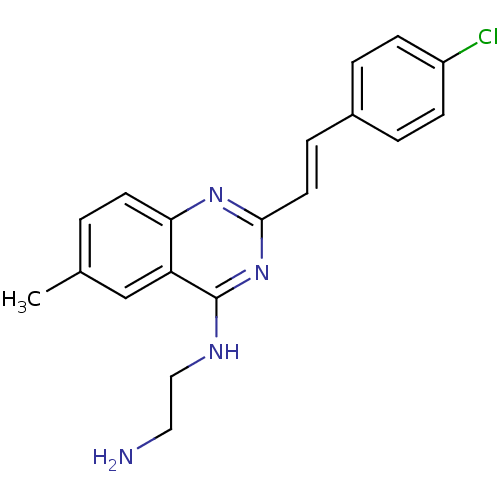
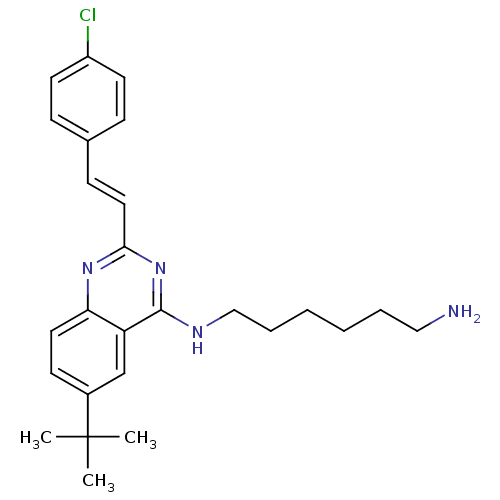
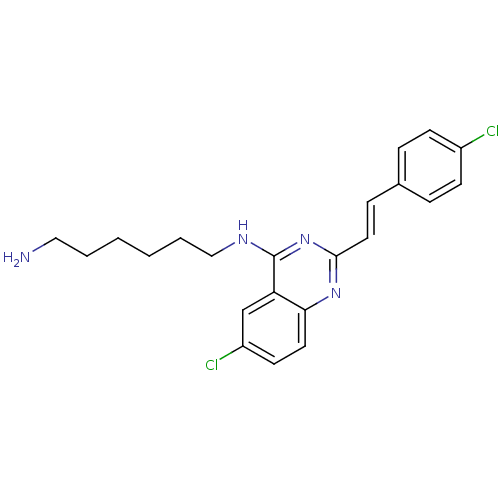
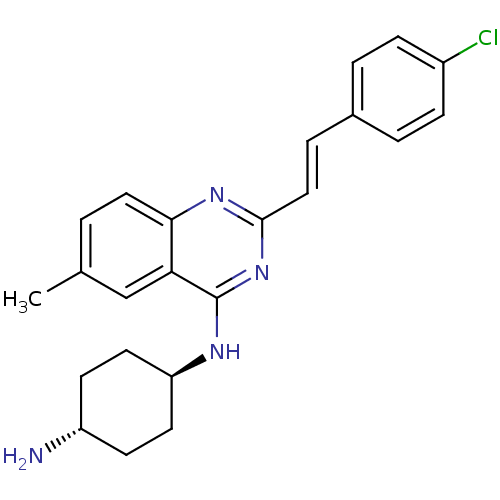
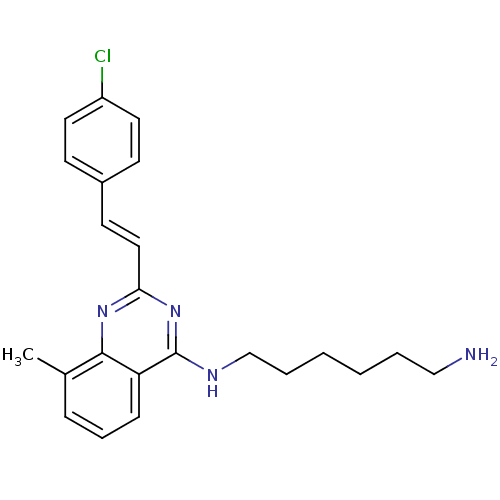
Report error Found 48 Enz. Inhib. hit(s) with all data for entry = 3019
Affinity DataKi: 1.40nM ΔG°: -50.5kJ/molepH: 7.8 T: 25°CAssay Description:Competitive binding displacement analysis was performed with membranes prepared from CHO-K1 cells stably expressing receptors. After incubation, samp...More data for this Ligand-Target Pair
Affinity DataKi: 2.10nM ΔG°: -49.5kJ/molepH: 7.8 T: 25°CAssay Description:Competitive binding displacement analysis was performed with membranes prepared from CHO-K1 cells stably expressing receptors. After incubation, samp...More data for this Ligand-Target Pair
Affinity DataKi: 43nM ΔG°: -42.0kJ/molepH: 7.8 T: 25°CAssay Description:Competitive binding displacement analysis was performed with membranes prepared from CHO-K1 cells stably expressing receptors. After incubation, samp...More data for this Ligand-Target Pair
Affinity DataKi: 60nMAssay Description:Competitive binding displacement analysis was performed with membranes prepared from CHO-K1 cells stably expressing receptors. After incubation, samp...More data for this Ligand-Target Pair
Affinity DataKi: 89nM ΔG°: -40.2kJ/molepH: 7.8 T: 25°CAssay Description:Competitive binding displacement analysis was performed with membranes prepared from CHO-K1 cells stably expressing receptors. After incubation, samp...More data for this Ligand-Target Pair
Affinity DataKi: 120nMAssay Description:Competitive binding displacement analysis was performed with membranes prepared from CHO-K1 cells stably expressing receptors. After incubation, samp...More data for this Ligand-Target Pair
Affinity DataKi: 133nM ΔG°: -39.2kJ/molepH: 7.8 T: 25°CAssay Description:Competitive binding displacement analysis was performed with membranes prepared from CHO-K1 cells stably expressing receptors. After incubation, samp...More data for this Ligand-Target Pair
Affinity DataKi: 140nM ΔG°: -39.1kJ/molepH: 7.8 T: 25°CAssay Description:Competitive binding displacement analysis was performed with membranes prepared from CHO-K1 cells stably expressing receptors. After incubation, samp...More data for this Ligand-Target Pair
Affinity DataKi: 150nMAssay Description:Competitive binding displacement analysis was performed with membranes prepared from CHO-K1 cells stably expressing receptors. After incubation, samp...More data for this Ligand-Target Pair
Affinity DataKi: 152nM ΔG°: -38.9kJ/molepH: 7.8 T: 25°CAssay Description:Competitive binding displacement analysis was performed with membranes prepared from CHO-K1 cells stably expressing receptors. After incubation, samp...More data for this Ligand-Target Pair
Affinity DataKi: 160nM ΔG°: -38.8kJ/molepH: 7.8 T: 25°CAssay Description:Competitive binding displacement analysis was performed with membranes prepared from CHO-K1 cells stably expressing receptors. After incubation, samp...More data for this Ligand-Target Pair
Affinity DataKi: 162nM ΔG°: -38.8kJ/molepH: 7.8 T: 25°CAssay Description:Competitive binding displacement analysis was performed with membranes prepared from CHO-K1 cells stably expressing receptors. After incubation, samp...More data for this Ligand-Target Pair
Affinity DataKi: 170nMAssay Description:Competitive binding displacement analysis was performed with membranes prepared from CHO-K1 cells stably expressing receptors. After incubation, samp...More data for this Ligand-Target Pair
Affinity DataKi: 180nMAssay Description:Competitive binding displacement analysis was performed with membranes prepared from CHO-K1 cells stably expressing receptors. After incubation, samp...More data for this Ligand-Target Pair
Affinity DataKi: 180nMAssay Description:Competitive binding displacement analysis was performed with membranes prepared from CHO-K1 cells stably expressing receptors. After incubation, samp...More data for this Ligand-Target Pair
Affinity DataKi: 194nM ΔG°: -38.3kJ/molepH: 7.8 T: 25°CAssay Description:Competitive binding displacement analysis was performed with membranes prepared from CHO-K1 cells stably expressing receptors. After incubation, samp...More data for this Ligand-Target Pair
Affinity DataKi: 200nMAssay Description:Competitive binding displacement analysis was performed with membranes prepared from CHO-K1 cells stably expressing receptors. After incubation, samp...More data for this Ligand-Target Pair
Affinity DataKi: 200nM ΔG°: -38.2kJ/molepH: 7.8 T: 25°CAssay Description:Competitive binding displacement analysis was performed with membranes prepared from CHO-K1 cells stably expressing receptors. After incubation, samp...More data for this Ligand-Target Pair
Affinity DataKi: 220nMAssay Description:Competitive binding displacement analysis was performed with membranes prepared from CHO-K1 cells stably expressing receptors. After incubation, samp...More data for this Ligand-Target Pair
Affinity DataKi: 232nM ΔG°: -37.9kJ/molepH: 7.8 T: 25°CAssay Description:Competitive binding displacement analysis was performed with membranes prepared from CHO-K1 cells stably expressing receptors. After incubation, samp...More data for this Ligand-Target Pair
Affinity DataKi: 240nMAssay Description:Competitive binding displacement analysis was performed with membranes prepared from CHO-K1 cells stably expressing receptors. After incubation, samp...More data for this Ligand-Target Pair
Affinity DataKi: 250nMAssay Description:Competitive binding displacement analysis was performed with membranes prepared from CHO-K1 cells stably expressing receptors. After incubation, samp...More data for this Ligand-Target Pair
Affinity DataKi: 270nMAssay Description:Competitive binding displacement analysis was performed with membranes prepared from CHO-K1 cells stably expressing receptors. After incubation, samp...More data for this Ligand-Target Pair
Affinity DataKi: 292nM ΔG°: -37.3kJ/molepH: 7.8 T: 25°CAssay Description:Competitive binding displacement analysis was performed with membranes prepared from CHO-K1 cells stably expressing receptors. After incubation, samp...More data for this Ligand-Target Pair
Affinity DataKi: 294nM ΔG°: -37.3kJ/molepH: 7.8 T: 25°CAssay Description:Competitive binding displacement analysis was performed with membranes prepared from CHO-K1 cells stably expressing receptors. After incubation, samp...More data for this Ligand-Target Pair
Affinity DataKi: 300nMAssay Description:Competitive binding displacement analysis was performed with membranes prepared from CHO-K1 cells stably expressing receptors. After incubation, samp...More data for this Ligand-Target Pair
Affinity DataKi: 300nMAssay Description:Competitive binding displacement analysis was performed with membranes prepared from CHO-K1 cells stably expressing receptors. After incubation, samp...More data for this Ligand-Target Pair
Affinity DataKi: 310nMAssay Description:Competitive binding displacement analysis was performed with membranes prepared from CHO-K1 cells stably expressing receptors. After incubation, samp...More data for this Ligand-Target Pair
Affinity DataKi: 330nMAssay Description:Competitive binding displacement analysis was performed with membranes prepared from CHO-K1 cells stably expressing receptors. After incubation, samp...More data for this Ligand-Target Pair
Affinity DataKi: 380nMAssay Description:Competitive binding displacement analysis was performed with membranes prepared from CHO-K1 cells stably expressing receptors. After incubation, samp...More data for this Ligand-Target Pair
Affinity DataKi: 380nMAssay Description:Competitive binding displacement analysis was performed with membranes prepared from CHO-K1 cells stably expressing receptors. After incubation, samp...More data for this Ligand-Target Pair
Affinity DataKi: 383nM ΔG°: -36.6kJ/molepH: 7.8 T: 25°CAssay Description:Competitive binding displacement analysis was performed with membranes prepared from CHO-K1 cells stably expressing receptors. After incubation, samp...More data for this Ligand-Target Pair
Affinity DataKi: 410nMAssay Description:Competitive binding displacement analysis was performed with membranes prepared from CHO-K1 cells stably expressing receptors. After incubation, samp...More data for this Ligand-Target Pair
Affinity DataKi: 410nMAssay Description:Competitive binding displacement analysis was performed with membranes prepared from CHO-K1 cells stably expressing receptors. After incubation, samp...More data for this Ligand-Target Pair
Affinity DataKi: 420nM ΔG°: -36.4kJ/molepH: 7.8 T: 25°CAssay Description:Competitive binding displacement analysis was performed with membranes prepared from CHO-K1 cells stably expressing receptors. After incubation, samp...More data for this Ligand-Target Pair
Affinity DataKi: 510nMAssay Description:Competitive binding displacement analysis was performed with membranes prepared from CHO-K1 cells stably expressing receptors. After incubation, samp...More data for this Ligand-Target Pair
Affinity DataKi: 538nM ΔG°: -35.8kJ/molepH: 7.8 T: 25°CAssay Description:Competitive binding displacement analysis was performed with membranes prepared from CHO-K1 cells stably expressing receptors. After incubation, samp...More data for this Ligand-Target Pair
Affinity DataKi: 653nM ΔG°: -35.3kJ/molepH: 7.8 T: 25°CAssay Description:Competitive binding displacement analysis was performed with membranes prepared from CHO-K1 cells stably expressing receptors. After incubation, samp...More data for this Ligand-Target Pair
Affinity DataKi: 795nM ΔG°: -34.8kJ/molepH: 7.8 T: 25°CAssay Description:Competitive binding displacement analysis was performed with membranes prepared from CHO-K1 cells stably expressing receptors. After incubation, samp...More data for this Ligand-Target Pair
Affinity DataKi: 808nM ΔG°: -34.8kJ/molepH: 7.8 T: 25°CAssay Description:Competitive binding displacement analysis was performed with membranes prepared from CHO-K1 cells stably expressing receptors. After incubation, samp...More data for this Ligand-Target Pair
Affinity DataKi: 930nMAssay Description:Competitive binding displacement analysis was performed with membranes prepared from CHO-K1 cells stably expressing receptors. After incubation, samp...More data for this Ligand-Target Pair
Affinity DataKi: 972nM ΔG°: -34.3kJ/molepH: 7.8 T: 25°CAssay Description:Competitive binding displacement analysis was performed with membranes prepared from CHO-K1 cells stably expressing receptors. After incubation, samp...More data for this Ligand-Target Pair
Affinity DataKi: 1.20E+3nMAssay Description:Competitive binding displacement analysis was performed with membranes prepared from CHO-K1 cells stably expressing receptors. After incubation, samp...More data for this Ligand-Target Pair
Affinity DataKi: 2.30E+3nMAssay Description:Competitive binding displacement analysis was performed with membranes prepared from CHO-K1 cells stably expressing receptors. After incubation, samp...More data for this Ligand-Target Pair
Affinity DataKi: 2.40E+3nMAssay Description:Competitive binding displacement analysis was performed with membranes prepared from CHO-K1 cells stably expressing receptors. After incubation, samp...More data for this Ligand-Target Pair
Affinity DataKi: 3.60E+3nMAssay Description:Competitive binding displacement analysis was performed with membranes prepared from CHO-K1 cells stably expressing receptors. After incubation, samp...More data for this Ligand-Target Pair
Affinity DataKi: 4.00E+3nMAssay Description:Competitive binding displacement analysis was performed with membranes prepared from CHO-K1 cells stably expressing receptors. After incubation, samp...More data for this Ligand-Target Pair
Affinity DataKi: 4.30E+3nMAssay Description:Competitive binding displacement analysis was performed with membranes prepared from CHO-K1 cells stably expressing receptors. After incubation, samp...More data for this Ligand-Target Pair