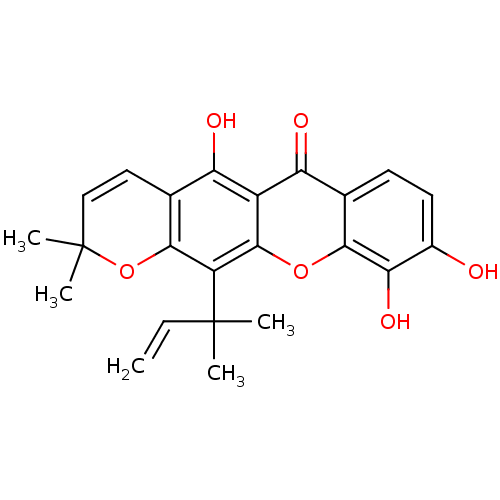
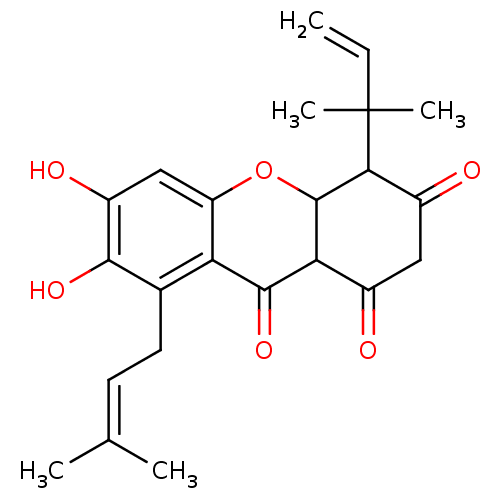
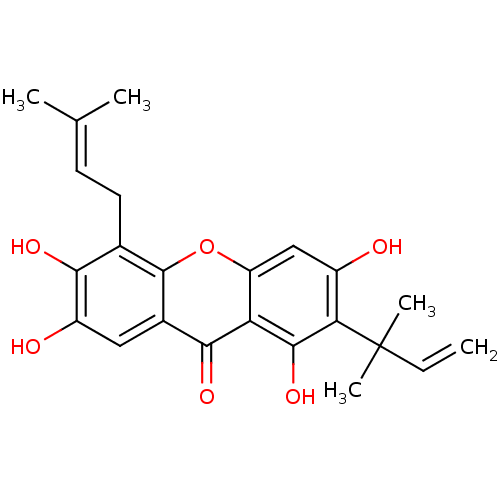
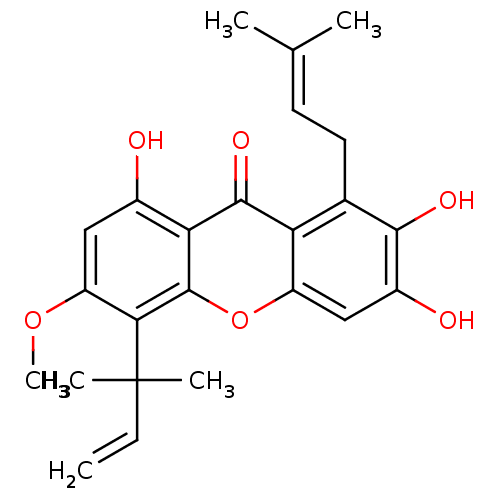
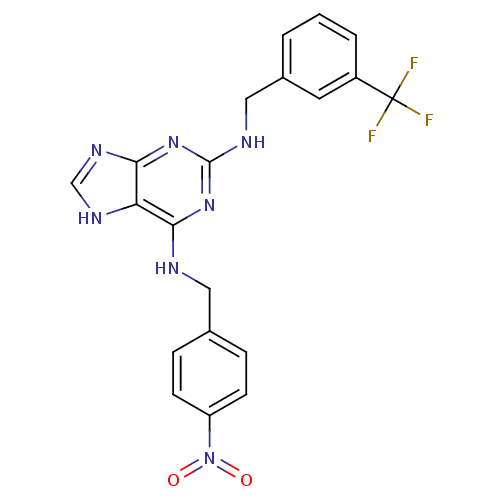
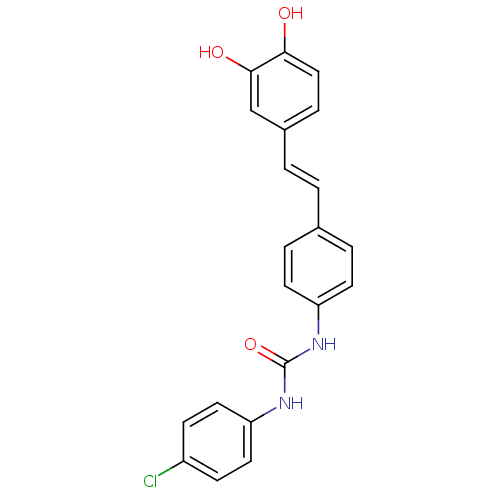
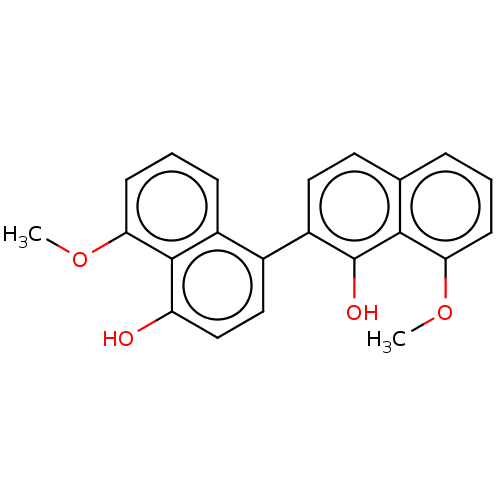
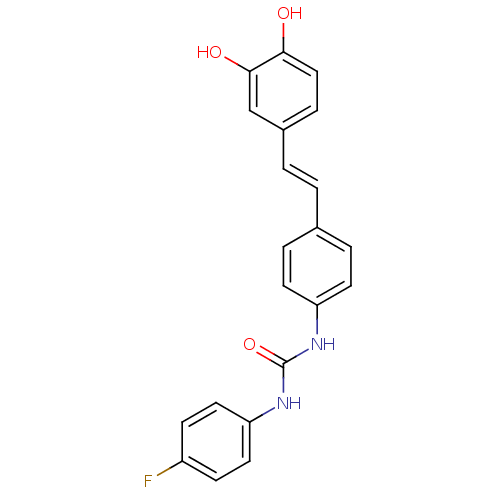
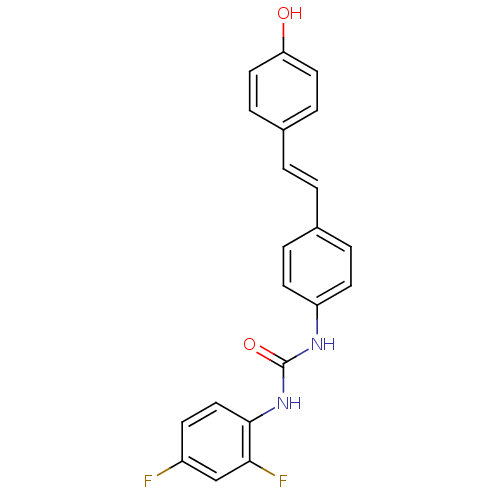
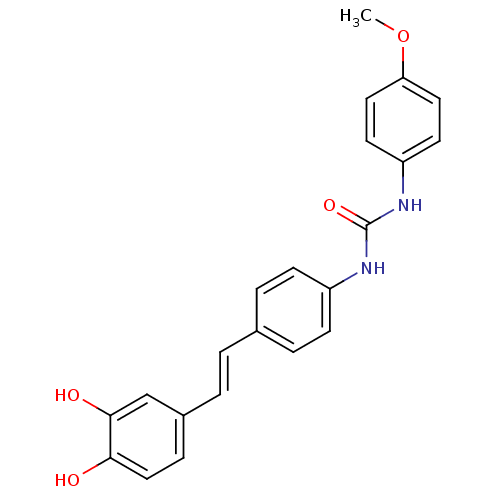
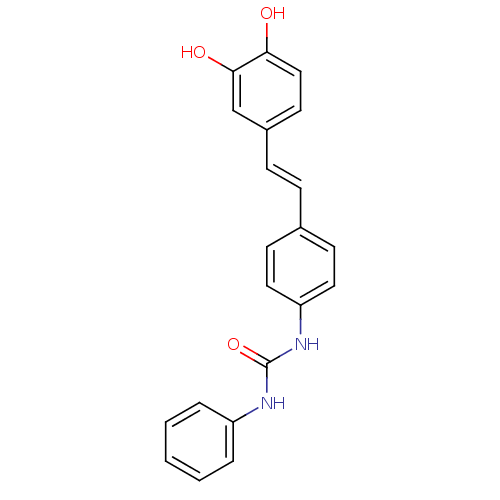
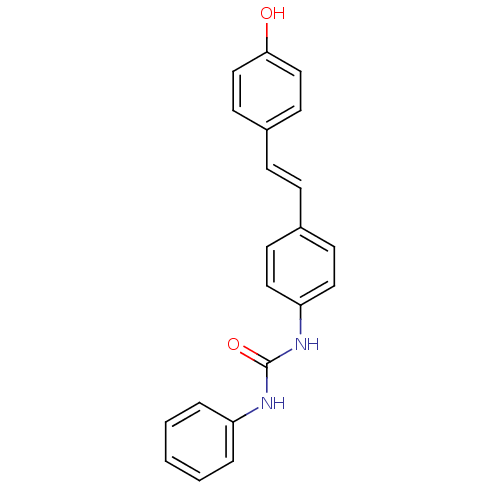
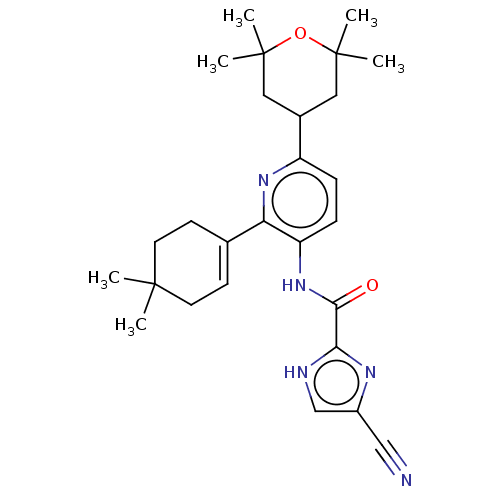
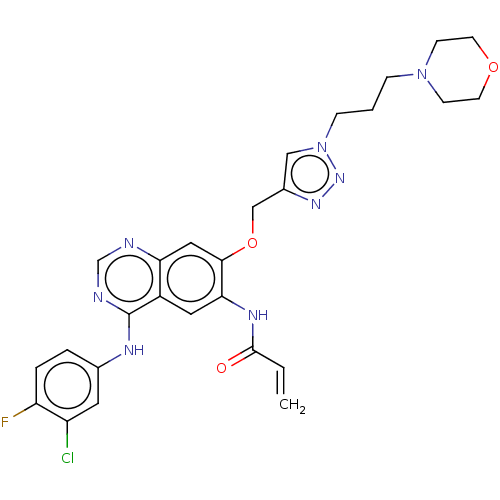
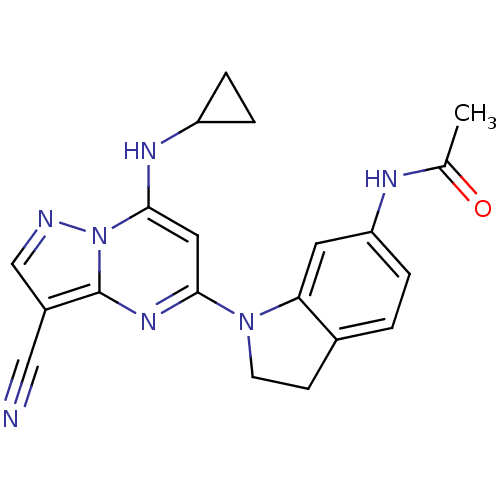
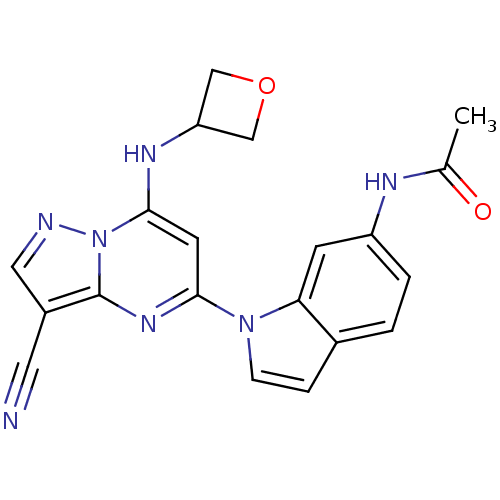
TargetPeroxisome proliferator-activated receptor delta(Homo sapiens (Human))
Seoul National University
Curated by ChEMBL
Seoul National University
Curated by ChEMBL
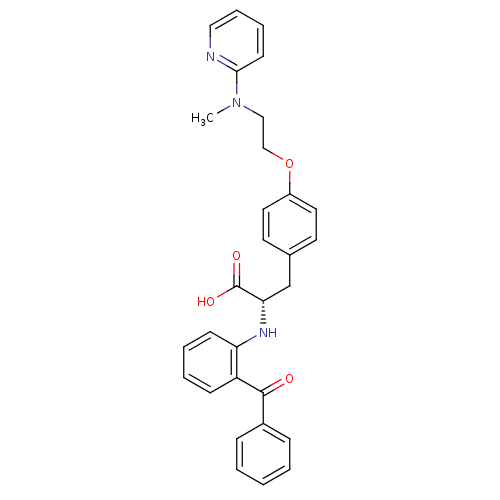
Affinity DataKi: 1nMAssay Description:Binding affinity to PPARdelta (unknown origin) assessed as inhibition constant by Cheng-Prusoff equation analysisMore data for this Ligand-Target Pair
TargetPeroxisome proliferator-activated receptor gamma(Homo sapiens (Human))
Seoul National University
Curated by ChEMBL
Seoul National University
Curated by ChEMBL
Affinity DataKi: 2nMAssay Description:Binding affinity to PPARgamma (unknown origin) assessed as inhibition constant by Cheng-Prusoff equation analysisMore data for this Ligand-Target Pair
TargetPeroxisome proliferator-activated receptor alpha(Homo sapiens (Human))
Seoul National University
Curated by ChEMBL
Seoul National University
Curated by ChEMBL
Affinity DataKi: 7nMAssay Description:Binding affinity to PPARalpha (unknown origin) assessed as inhibition constant by Cheng-Prusoff equation analysisMore data for this Ligand-Target Pair
TargetSialidase(Clostridium perfringens)
Graduate School Of Gyeongsang National University
Curated by ChEMBL
Graduate School Of Gyeongsang National University
Curated by ChEMBL
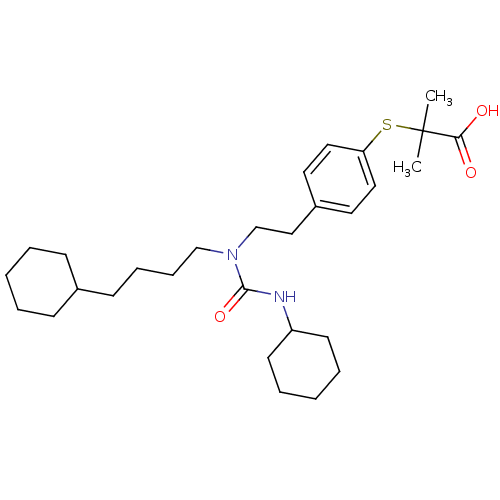
Affinity DataKi: 58nMAssay Description:Competitive inhibition of Clostridium perfringens neuraminidase by Lineweaver-Burke plot and Dixon plotMore data for this Ligand-Target Pair
TargetSialidase(Clostridium perfringens)
Graduate School Of Gyeongsang National University
Curated by ChEMBL
Graduate School Of Gyeongsang National University
Curated by ChEMBL
Affinity DataKi: 98nMAssay Description:Competitive inhibition of Clostridium perfringens neuraminidase by Lineweaver-Burke plot and Dixon plotMore data for this Ligand-Target Pair
TargetSialidase(Clostridium perfringens)
Graduate School Of Gyeongsang National University
Curated by ChEMBL
Graduate School Of Gyeongsang National University
Curated by ChEMBL
Affinity DataKi: 103nMAssay Description:Competitive inhibition of Clostridium perfringens neuraminidase by Lineweaver-Burke plot and Dixon plotMore data for this Ligand-Target Pair
TargetSialidase(Clostridium perfringens)
Graduate School Of Gyeongsang National University
Curated by ChEMBL
Graduate School Of Gyeongsang National University
Curated by ChEMBL
Affinity DataKi: 127nMAssay Description:Apparent binding affinity at Clostridium perfringens neuraminidase by fluorimetryMore data for this Ligand-Target Pair
TargetSialidase(Clostridium perfringens)
Graduate School Of Gyeongsang National University
Curated by ChEMBL
Graduate School Of Gyeongsang National University
Curated by ChEMBL
Affinity DataKi: 136nMAssay Description:Competitive inhibition of Clostridium perfringens neuraminidase by Lineweaver-Burke plot and Dixon plotMore data for this Ligand-Target Pair
TargetSialidase(Clostridium perfringens)
Graduate School Of Gyeongsang National University
Curated by ChEMBL
Graduate School Of Gyeongsang National University
Curated by ChEMBL
Affinity DataKi: 138nMAssay Description:Competitive inhibition of Clostridium perfringens neuraminidase by Lineweaver-Burke plot and Dixon plotMore data for this Ligand-Target Pair
TargetSialidase(Clostridium perfringens)
Graduate School Of Gyeongsang National University
Curated by ChEMBL
Graduate School Of Gyeongsang National University
Curated by ChEMBL
Affinity DataKi: 143nMAssay Description:Competitive inhibition of Clostridium perfringens neuraminidase by Lineweaver-Burke plot and Dixon plotMore data for this Ligand-Target Pair
TargetSialidase(Clostridium perfringens)
Graduate School Of Gyeongsang National University
Curated by ChEMBL
Graduate School Of Gyeongsang National University
Curated by ChEMBL
Affinity DataKi: 214nMAssay Description:Apparent binding affinity at Clostridium perfringens neuraminidase by fluorimetryMore data for this Ligand-Target Pair
TargetSialidase(Clostridium perfringens)
Graduate School Of Gyeongsang National University
Curated by ChEMBL
Graduate School Of Gyeongsang National University
Curated by ChEMBL
Affinity DataKi: 436nMAssay Description:Apparent binding affinity at Clostridium perfringens neuraminidase by fluorimetryMore data for this Ligand-Target Pair
TargetSialidase(Clostridium perfringens)
Graduate School Of Gyeongsang National University
Curated by ChEMBL
Graduate School Of Gyeongsang National University
Curated by ChEMBL
Affinity DataKi: 477nMAssay Description:Apparent binding affinity at Clostridium perfringens neuraminidase by fluorimetryMore data for this Ligand-Target Pair
TargetSialidase(Clostridium perfringens)
Graduate School Of Gyeongsang National University
Curated by ChEMBL
Graduate School Of Gyeongsang National University
Curated by ChEMBL
Affinity DataKi: 510nMAssay Description:Apparent binding affinity at Clostridium perfringens neuraminidase by fluorimetryMore data for this Ligand-Target Pair
TargetSialidase(Clostridium perfringens)
Graduate School Of Gyeongsang National University
Curated by ChEMBL
Graduate School Of Gyeongsang National University
Curated by ChEMBL
Affinity DataKi: 568nMAssay Description:Apparent binding affinity at Clostridium perfringens neuraminidase by fluorimetryMore data for this Ligand-Target Pair
TargetPeroxisome proliferator-activated receptor gamma(Homo sapiens (Human))
Seoul National University
Curated by ChEMBL
Seoul National University
Curated by ChEMBL
Affinity DataKi: 600nMAssay Description:Binding affinity to PPARgamma (unknown origin) assessed as inhibition constant by Cheng-Prusoff equation analysisMore data for this Ligand-Target Pair
TargetSialidase(Clostridium perfringens)
Graduate School Of Gyeongsang National University
Curated by ChEMBL
Graduate School Of Gyeongsang National University
Curated by ChEMBL
Affinity DataKi: 950nMAssay Description:Competitive inhibition of Clostridium perfringens neuraminidase by Lineweaver-Burke plot and Dixon plotMore data for this Ligand-Target Pair
TargetSialidase(Clostridium perfringens)
Graduate School Of Gyeongsang National University
Curated by ChEMBL
Graduate School Of Gyeongsang National University
Curated by ChEMBL
Affinity DataKi: 2.23E+3nMAssay Description:Apparent binding affinity at Clostridium perfringens neuraminidase by fluorimetryMore data for this Ligand-Target Pair
Affinity DataKi: 3.20E+3nM IC50: 8.40E+3nMAssay Description:All enzymatic activities were determined by using the appropriate substrate (p-nitrophenyl-alpha-D-glucopyranoside, p-nitrophenyl-beta-D-gulcopyranos...More data for this Ligand-Target Pair
Affinity DataKi: 4.30E+3nM ΔG°: -31.9kJ/mole IC50: 1.02E+4nMpH: 8.0 T: 2°CAssay Description:IP3K reactions were carried out in a 100ul solution that contained Tris-Cl, EGTA, ATP, DTT, 2,3-diphosphoglycerate, D-l(1,4,5)P3, [3H]-l(1,4,5)P3 and...More data for this Ligand-Target Pair
Affinity DataKi: 4.60E+3nM IC50: 1.43E+4nMAssay Description:All enzymatic activities were determined by using the appropriate substrate (p-nitrophenyl-alpha-D-glucopyranoside, p-nitrophenyl-beta-D-gulcopyranos...More data for this Ligand-Target Pair
TargetPeroxisome proliferator-activated receptor gamma(Homo sapiens (Human))
Seoul National University
Curated by ChEMBL
Seoul National University
Curated by ChEMBL
Affinity DataKi: 6.60E+3nMAssay Description:Binding affinity to PPARgamma (unknown origin) assessed as inhibition constant by Cheng-Prusoff equation analysisMore data for this Ligand-Target Pair
Affinity DataKi: 7.20E+3nM IC50: 1.98E+4nMAssay Description:All enzymatic activities were determined by using the appropriate substrate (p-nitrophenyl-alpha-D-glucopyranoside, p-nitrophenyl-beta-D-gulcopyranos...More data for this Ligand-Target Pair
Affinity DataKi: 9.40E+3nM IC50: 1.85E+4nMAssay Description:All enzymatic activities were determined by using the appropriate substrate (p-nitrophenyl-alpha-D-glucopyranoside, p-nitrophenyl-beta-D-gulcopyranos...More data for this Ligand-Target Pair
TargetPeroxisome proliferator-activated receptor alpha(Homo sapiens (Human))
Seoul National University
Curated by ChEMBL
Seoul National University
Curated by ChEMBL
Affinity DataKi: 9.40E+3nMAssay Description:Binding affinity to PPARalpha (unknown origin) assessed as inhibition constant by Cheng-Prusoff equation analysisMore data for this Ligand-Target Pair
Affinity DataKi: 1.05E+4nM IC50: 1.95E+4nMAssay Description:All enzymatic activities were determined by using the appropriate substrate (p-nitrophenyl-alpha-D-glucopyranoside, p-nitrophenyl-beta-D-gulcopyranos...More data for this Ligand-Target Pair
Affinity DataKi: 1.06E+4nM IC50: 4.21E+4nMAssay Description:All enzymatic activities were determined by using the appropriate substrate (p-nitrophenyl-alpha-D-glucopyranoside, p-nitrophenyl-beta-D-gulcopyranos...More data for this Ligand-Target Pair
Affinity DataKi: 1.21E+4nM IC50: 2.88E+4nMAssay Description:All enzymatic activities were determined by using the appropriate substrate (p-nitrophenyl-alpha-D-glucopyranoside, p-nitrophenyl-beta-D-gulcopyranos...More data for this Ligand-Target Pair
Affinity DataKi: 1.70E+4nM IC50: 2.91E+4nMAssay Description:All enzymatic activities were determined by using the appropriate substrate (p-nitrophenyl-alpha-D-glucopyranoside, p-nitrophenyl-beta-D-gulcopyranos...More data for this Ligand-Target Pair
Affinity DataKi: 1.80E+4nM IC50: 3.95E+4nMAssay Description:All enzymatic activities were determined by using the appropriate substrate (p-nitrophenyl-alpha-D-glucopyranoside, p-nitrophenyl-beta-D-gulcopyranos...More data for this Ligand-Target Pair
TargetPeroxisome proliferator-activated receptor delta(Homo sapiens (Human))
Seoul National University
Curated by ChEMBL
Seoul National University
Curated by ChEMBL
Affinity DataKi: 1.88E+4nMAssay Description:Binding affinity to PPARdelta (unknown origin) assessed as inhibition constant by Cheng-Prusoff equation analysisMore data for this Ligand-Target Pair
TargetPeroxisome proliferator-activated receptor gamma(Homo sapiens (Human))
Seoul National University
Curated by ChEMBL
Seoul National University
Curated by ChEMBL
Affinity DataKi: 1.91E+4nMAssay Description:Binding affinity to PPARgamma (unknown origin) assessed as inhibition constant by Cheng-Prusoff equation analysisMore data for this Ligand-Target Pair
Affinity DataKi: 2.15E+4nM IC50: 2.64E+4nMAssay Description:All enzymatic activities were determined by using the appropriate substrate (p-nitrophenyl-alpha-D-glucopyranoside, p-nitrophenyl-beta-D-gulcopyranos...More data for this Ligand-Target Pair
Affinity DataKi: 3.51E+4nM IC50: 4.64E+4nMAssay Description:All enzymatic activities were determined by using the appropriate substrate (p-nitrophenyl-alpha-D-glucopyranoside, p-nitrophenyl-beta-D-gulcopyranos...More data for this Ligand-Target Pair
TargetMacrophage colony-stimulating factor 1 receptor(Homo sapiens (Human))
Chung-Ang University
Curated by ChEMBL
Chung-Ang University
Curated by ChEMBL
Affinity DataIC50: 1.20nMAssay Description:Inhibition of CSF1R (unknown origin)More data for this Ligand-Target Pair
TargetCholesteryl ester transfer protein(Homo sapiens (Human))
Chong Kun Dang Pharmaceutical
US Patent
Chong Kun Dang Pharmaceutical
US Patent
Affinity DataIC50: 1.5nMAssay Description:As a protein source for cholesteryl ester transfer, plasma from healthy persons was used, and as a cholesteryl ester receptor, LDL from healthy perso...More data for this Ligand-Target Pair
TargetCholesteryl ester transfer protein(Homo sapiens (Human))
Chong Kun Dang Pharmaceutical
US Patent
Chong Kun Dang Pharmaceutical
US Patent
Affinity DataIC50: 1.60nMAssay Description:As a protein source for cholesteryl ester transfer, plasma from healthy persons was used, and as a cholesteryl ester receptor, LDL from healthy perso...More data for this Ligand-Target Pair
TargetCholesteryl ester transfer protein(Homo sapiens (Human))
Chong Kun Dang Pharmaceutical
US Patent
Chong Kun Dang Pharmaceutical
US Patent
Affinity DataIC50: 1.90nMAssay Description:As a protein source for cholesteryl ester transfer, plasma from healthy persons was used, and as a cholesteryl ester receptor, LDL from healthy perso...More data for this Ligand-Target Pair
TargetCholesteryl ester transfer protein(Homo sapiens (Human))
Chong Kun Dang Pharmaceutical
US Patent
Chong Kun Dang Pharmaceutical
US Patent
Affinity DataIC50: 1.90nMAssay Description:As a protein source for cholesteryl ester transfer, plasma from healthy persons was used, and as a cholesteryl ester receptor, LDL from healthy perso...More data for this Ligand-Target Pair
TargetCholesteryl ester transfer protein(Homo sapiens (Human))
Chong Kun Dang Pharmaceutical
US Patent
Chong Kun Dang Pharmaceutical
US Patent
Affinity DataIC50: 2.5nMAssay Description:As a protein source for cholesteryl ester transfer, plasma from healthy persons was used, and as a cholesteryl ester receptor, LDL from healthy perso...More data for this Ligand-Target Pair
TargetMacrophage colony-stimulating factor 1 receptor(Homo sapiens (Human))
Chung-Ang University
Curated by ChEMBL
Chung-Ang University
Curated by ChEMBL
Affinity DataIC50: 3.20nMAssay Description:Inhibition of CSF1R (unknown origin) (538 to 972 residues) expressed in baculovirus expression system using SYEGNSYTFIDPTQ as substrate incubated for...More data for this Ligand-Target Pair
TargetCholesteryl ester transfer protein(Homo sapiens (Human))
Chong Kun Dang Pharmaceutical
US Patent
Chong Kun Dang Pharmaceutical
US Patent
Affinity DataIC50: 3.90nMAssay Description:As a protein source for cholesteryl ester transfer, plasma from healthy persons was used, and as a cholesteryl ester receptor, LDL from healthy perso...More data for this Ligand-Target Pair
Affinity DataIC50: 4nMAssay Description:Inhibition of human N-terminal GST-tagged EGFR T790M/L858R double mutant (696 to end residues) expressed in baculovirus infected sf21 cells using pol...More data for this Ligand-Target Pair
Affinity DataIC50: 4nMAssay Description:Inhibition of full length N-terminus His-tagged human CK2alpha expressed in insect Sf21 cells using BODIPY-FL-RRRDDDSDDD-CONH2 as substrate after 20 ...More data for this Ligand-Target Pair
Affinity DataIC50: 4nMAssay Description:Inhibition of full length N-terminus His-tagged human CK2alpha expressed in insect Sf21 cells using BODIPY-FL-RRRDDDSDDD-CONH2 as substrate after 20 ...More data for this Ligand-Target Pair
Affinity DataIC50: 4nMAssay Description:Inhibition of human N-terminal GST-tagged EGFR T790M/L858R double mutant (696 to end residues) expressed in baculovirus infected sf21 cells using pol...More data for this Ligand-Target Pair
Affinity DataIC50: 4nMAssay Description:Inhibition of human N-terminal GST-tagged EGFR T790M/L858R double mutant (696 to end residues) expressed in baculovirus infected sf21 cells using pol...More data for this Ligand-Target Pair
TargetVascular endothelial growth factor receptor 2(Homo sapiens (Human))
Incheon University Industry Academic Cooperation Foundation
US Patent
Incheon University Industry Academic Cooperation Foundation
US Patent
Affinity DataIC50: 4.10nMAssay Description:The inhibitory activities of the compounds of the present invention against VEGFR-2 tyrosine kinase were analyzed using ADP-Glo™ kinase assay kit com...More data for this Ligand-Target Pair
TargetCholesteryl ester transfer protein(Homo sapiens (Human))
Chong Kun Dang Pharmaceutical
US Patent
Chong Kun Dang Pharmaceutical
US Patent
Affinity DataIC50: 5nMAssay Description:As a protein source for cholesteryl ester transfer, plasma from healthy persons was used, and as a cholesteryl ester receptor, LDL from healthy perso...More data for this Ligand-Target Pair
Affinity DataIC50: 5nMAssay Description:Inhibition of human N-terminal GST-tagged EGFR T790M/L858R double mutant (696 to end residues) expressed in baculovirus infected sf21 cells using pol...More data for this Ligand-Target Pair

 3D Structure (crystal)
3D Structure (crystal)